Mar 9 2017
Like DNA, ribonucleic acid (RNA) is a type of polymeric biomolecule essential for life, playing important roles in gene processing. Short lengths of RNA called microRNA are more stable than longer RNA chains, and are found in common bodily fluids. The level of microRNA in bodily fluids is strongly correlated with the presence and advance of cancer. This means that microRNA can act as an easily accessible biomarker to diagnose cancer, which causes over 14% of deaths annually worldwide.
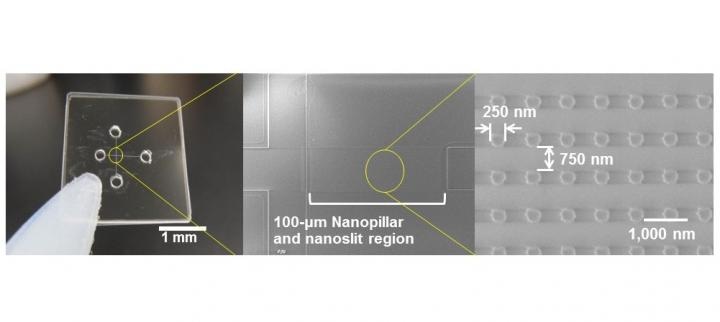 Quartz-made Nanopillars of 250-nm diameter were arrayed inside nanoslit region of 100-nm high and applied for ultrafast microRNA extraction from nucleic acids mixture. (Credit: Noritada Kaji)
Quartz-made Nanopillars of 250-nm diameter were arrayed inside nanoslit region of 100-nm high and applied for ultrafast microRNA extraction from nucleic acids mixture. (Credit: Noritada Kaji)
To use microRNA as a biomarker for cancer, it needs to be isolated by a rapid, efficient process. A collaboration led by researchers at Nagoya University has developed an innovative nanobiodevice that can separate microRNA from DNA/RNA mixtures obtained from cells in less than 100 ms.
The nanobiodevice consists of a quartz substrate containing a 25×100 μm array of "nanopillars" (small columns with a diameter of 250 nm and height of 100 nm) in shallow "nanoslits" with a height of 100 nm and fabricated in a microchannel by electron beam lithography.
The ability of the nanobiodevice to separate microRNA from DNA was first investigated using mixtures containing components with known concentrations. The team optimized the separation conditions, achieving almost complete separation of microRNA from DNA in just 20 ms. This is the fastest complete separation of microRNA to date.
The researchers then separated a mixture of microRNA, RNA, and DNA isolated from cells using the nanobiodevice. Separation with high resolution was realized in 100 ms. The nanobiodevice separated microRNA from RNA and DNA because of the different mobilities of these materials through the nanopillar region of the microchannel.
"We believe that the nanobiodevice separates microRNA from mixtures through a combination of two different physical behaviors of confined polymers in the nanopoillar array, non-equilibrium transport and entropic trapping," corresponding author Noritada Kaji says. "The applied electric field combines with the unique nanostructure of the nanobiodevice to generate a strong electric force that induces rapid concentration and separation."
The speed at which this nanobiodevice can separate microRNA from complex mixtures means that it is promising for integration with nanopore DNA sequencing, which aims to realize direct sequencing of DNA or RNA at a rate of 1 base/ms. The developed nanobiodevice separation approach may lead to faster, more reliable isolation of microRNA, facilitating its use as a biomarker to allow quicker and easier detection of cancer.
This study was conducted by Nagoya University, Kyushu University, Hokkaido University, and Osaka University.