Over the past few years, nanomaterials altered with organic molecules have been applied in a wide range of technologies such as smart nanomaterials, nanomedicine, and biosensing. An organic monolayer can be used to alter the nanoparticles. Typically the monolayer has functional groups that behave as hooks onto which organic molecules, including biomolecules, can attach covalently.
The surface coating of nanoparticles with these organic ligands can offer discrete properties such as improved solubility, stability, and wettability of the particles. For instance, a polar surface coating provides high aqueous solubility and eliminates the aggregation of nanoparticles.
With regard to biological applications, the polyethylene glycol polymer is usually used for coating nanoparticles as it improves their circulation time in the body, and they also experience less non-specific interactions as opposed to the unchanged nanoparticles.
The characterization of customized nanomaterials is necessary to enhance process efficiency and quality control evaluation. While a variety of spectroscopy methods, such as X-ray photoelectron spectroscopy and infrared spectroscopy, have been employed for the surface characterization of nanomaterials, they do not have the specificity in identification, as opposed to analytical technologies utilizing precise and high resolution mass spectrometry (MS).
Other techniques such as matrix assisted laser desorption (MALDI) Time-of-Flight (TOF) mass spectrometry technology, which offer mass precision and high resolution, have also been utilized. Conversely, the MALDI-TOF MS is restricted by an appropriate selection of matrix to test organic capped nanomaterials, matrix interference at the low masses (<300 Da), and uneven distribution of sample in matrix.
In this analysis, the use of ambient ionization TOF mass spectrometry is studied. It is a method that ionizes analytes in ambient conditions, to differentiate nanomaterials’ surface.
The Direct Sample Analysis (DSA™) system was used, which is an ambient ionization source using a “field free” corona discharge electron Reagent Ion Generator (RIG). The nanoparticles that were modified were layered on a steel mesh and the same are exposed to the source fixed at 350°C for just a few seconds.
Due to thermo-lability of the covalent bonds, the organic capped ligands are discharged from the nanoparticles. The source’s temperature is not adequate enough to volatilize the particles’ metallic core. Ionized by the DSA source, the emitted organic molecules subsequently pass the mass spectrometer and are identified with a high-resolution TOF MS providing a mass precision of ≤2 ppm determined at m/z 1000.
The existence of various types of ligands bound to nanoparticles was verified by utilizing the precise mass and isotope profile data provided by the TOF together with powerful visualization software tools. The DSA-TOF-MS offers a substitute technology for fast analysis of organically capped nanoparticles without any sample preparation.
Experiment
All nanomaterials were obtained from a single manufacturer, Nanocomposix. For this research, the PerkinElmer DSA source was utilized as the ambient source The DSA was placed on a PerkinElmer AxION™ 2 Time of Flight (TOF) mass spectrometer, and the DSA source was sustained at 350°C temperature during analysis. The DSA controller software™ was used to acquire mass spectra, and for data analysis the AxION Solo™ software was utilized. The samples were located on the mesh for analysis. For each sample, the total amount of nanomaterial loaded on the mesh was between 2 and 5 ug.
Results
Gold (Au) nanomaterials capped with frequently used organic ligands such as Cetyl trimethylammonium bromide (CTAB) and lipoic acid t were examined. Such ligands can then act as linkers for other types of molecules. Gold nanoparticles modified with lipoic acid were produced by decreasing the disulphide bond in lipoic acid with dithiothreitol before attaching with Au nanoparticles. In the reduced lipoic acid, the free thiol groups stick to the gold surface, leading to a powerful covalent bond. On testing the lipoic acid Au nanoparticles in negative mode, the [M-H]- ion of lipoic acid was obtained, signifying that the lipoic acid was discharged from the particles owing to thermolability of the thiol-Au linkage (Figure. 1).
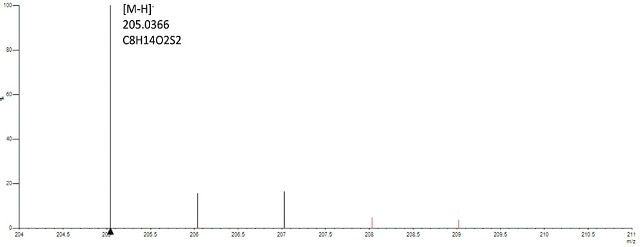
Figure 1. Spectrum of gold-lipoic acid nanoparticles analyzed in negative mode confirms presence of lipoic acid based on accurate mass (<2ppm) of the monoisptopic mass and isotope profile (<6%of theoritical).
The [M- H]- ion displayed mass precision within 1 ppm and isotope profile within 2% of the predictable value of lipoic acid, substantiating the existence of the ligand. Due to the oxidizing environment of the DSA source, the discharged lipoic acid is in its oxidized form.
On examining the CTAB-modified gold particles in positive mode, the [M+H]+ ion and the ion ensuing from loss of a methyl group [M-CH3+H]+ was displayed (Figure 2).
![Spectrum of gold-CTAB acid nanoparticles analyzed in positve mode shows the [M+H]+ ion corresponding to CTAB moiety and the ion showing loss of a methyl group from CTAB [M+H-CH3] +.](https://www.azonano.com/images/Article_Images/ImageForArticle_4229_44748320566122689720.jpg)
Figure 2. Spectrum of gold-CTAB acid nanoparticles analyzed in positve mode shows the [M+H]+ ion corresponding to CTAB moiety and the ion showing loss of a methyl group from CTAB [M+H-CH3] +.
The DSA-TOF was used to analyze the standard solution of branched polyethyleneimine (BPEI) used to alter the gold nanomaterials. The BPEI’s spectrum was typical of a polymer comprising repeating units (Figure 3).
![Spectrum of standard BPEI polymer solution shows repeating unit of 43.042 units corresponding to the elemental composition of [CH2CH2NH]n.](https://www.azonano.com/images/Article_Images/ImageForArticle_4229_44748320567314814444.jpg)
Figure 3. Spectrum of standard BPEI polymer solution shows repeating unit of 43.042 units corresponding to the elemental composition of [CH2CH2NH]n.
The repeating units varied by 43.042 Da relative to the elemental composition of [CH2CH2NH], validating the existence of BPEI. Lipoic acid was used as the linker to alter the nanoparticles with BPEI. Gold was attached covalently to the lipoic acid and the terminal amine group of BPEI and the free carboxyl group of lipoic acid was linked together, leading to an amide bond (Figure 4).
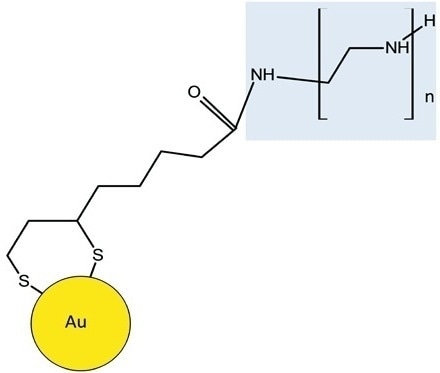
Figure 4. Schematic shows BPEI is attached to the lipoic acid-Au through an amide linkage. The BPEI moiety of the molecule is highlighted in blue.
When analyzed by DSA-TOF, the Au modified BPEI resulted in a spectrum analogous to the standard BPEI (Figure 5).
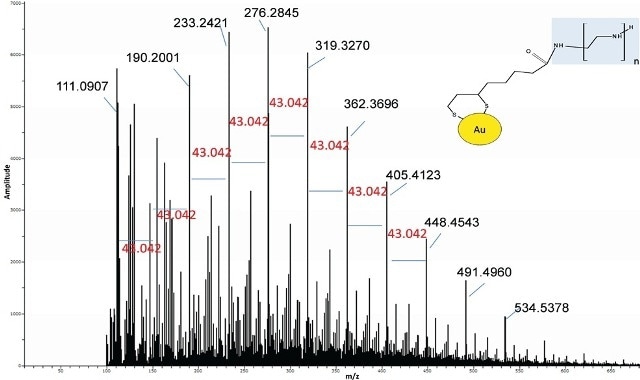
Figure 5. Spectrum of Au-lipoic acid-BPEI nanoparticles was similar to the BPEI standard suggesting BPEI was bound to the Au.
Nevertheless, a thorough examination of the spectrum also revealed ions related to lipoic acid-BPEI conjugate ions. In the modified gold nanomaterials, the extracted ion chromatograms (EICs) of some of the key BPEI ions conjugated to lipoic acid were seen but the same were missing in the Au particles and the control BPEI standard (Figure 6).
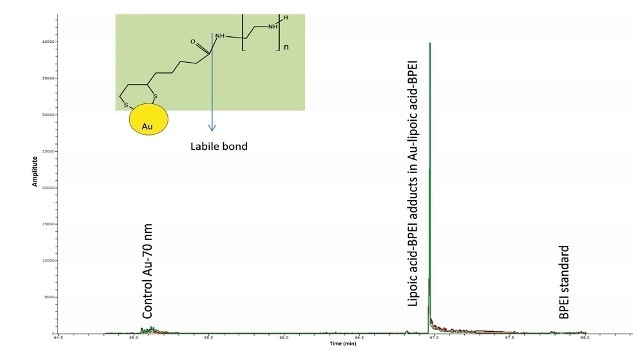
Figure 6. Overlay of EICs of m/z 421.2777, 464.3199, 507.3621 corresponding to the conjugates of the predominant ions of BPEI (m/z 233, 276, 319) with lipoic acid (minus water) were observed only in the Au-lipoic acid-BPEI sample and not in the negative controls (Au particles and BPEI standard). This suggests that BPEI is bound to AU via lipoic acid.
Amide bonds are thermo-labile in nature such as the one associating BPEI to lipoic acid. This may explain the cause for BPEI being the major ions in the mass spectra and not the BPEI-lipoic acid conjugates.
The DSA-TOF offers fast analysis without involving any widespread sample preparation,` which makes it an ideal method for rapid quality control evaluation of the altered nanomaterials.
Figure 7 shows the spectra obtained for BPEI conjugated gold nanoparticles from two different lots. The spectra from Lot 2 shows the classic BPEI polymer pattern unlike Lot 1 suggesting Lot 2 to be successfully modified by the ligand.
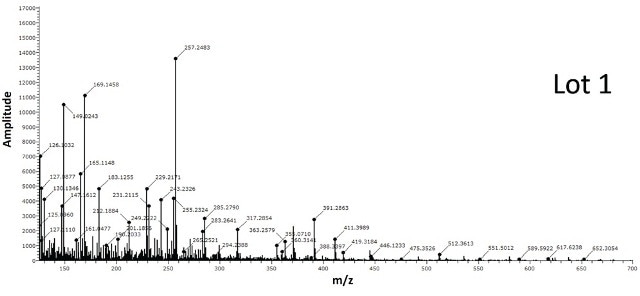
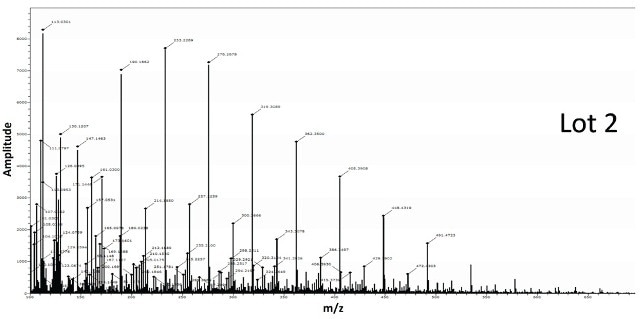
Figure 7. Two different lots of BPEI-lipoic acid-Au nanomaterials were analyzed by DSA-TOF (2 μg of each sample). Lot 2 showed the entire mass spectral profile of BPEI polymer, unlike Lot 1, suggesting it was successfully modified by BPEI
The spectra acquired for BPEI conjugated gold nanoparticles from two different lots is shown in Figure 7. The spectra from Lot 2 displays the standard BPEI polymer pattern unlike Lot 1, indicating Lot 2 to be effectively altered by the ligand.
In addition to precise mass and isotope profile data rendered by the TOF mass spectrometer, powerful software visualization tools employing the AxION Solo software can assist in detecting the analytes of interest in bulk batches of samples or in nanoparticles comprising blends of capped ligands.
To explain this clearly, the green color coding shown in Figure 8 illustrates the existence of dodecanethiol dimer in the dodecanethiol capped gold nanoparticles and in the blend of nanoparticles comprising CTAB as well as dodecanethiol conjugated to the gold surface, but not in the negative controls color indicated in grey.
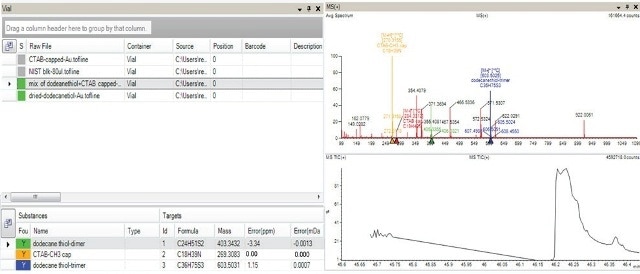
Figure 8. AxION Solo software shows presence of dodecanethiol (dimer and trimers) indicated in green color in the sample containing mixture of Au capped with dodecanethiol and Au capped with CTAB and also in the sample containing Au capped with dodecanethiol (top left hand corner of panel). The absence of the target in the negative controls (Au particles and Au capped with CTAB) is indicated in grey. The bottom left hand corner of the panel indicates all the target analytes along with the mass accuracy information for the selected sample which contained the mixture of capped ligands. The spectra for this mixed sample is shown in the top right hand corner.
Conclusion
The DSA-TOF serves as a quick analysis instrument for quality evaluation of the characterization of organic capped nanomaterials. Using exact mass and isotope profile data given by the TOF, coupled with powerful visualization software tools, the presence of the different types of ligands bound to the nanoparticles was confirmed.
In addition to simply detecting and validating a single type of organic monolayer bound covalently to the nanoparticle, bilayers were also identified wherein a second type of organic ligand was used to covalently modify a single organic monolayer.

This information has been sourced, reviewed and adapted from materials provided by PerkinElmer Inc.
For more information on this source, please visit PerkinElmer Inc.