Nanostructures are widely utilized in therapeutic approaches. New research, published in Informatics in Medicine Unlocked employs perovskite nanostructures as a suppression agent for COVID-19 for the first time
Image Credit: peterschreiber.media/Shutterstock.com
The COVID-19 global pandemic, caused by SARS-CoV-2, or coronavirus, started in early 2020 and became dominant in many countries. Millions of people are affected by COVID-19 globally, with pathologies varying from mild respiratory symptoms to acute respiratory distress syndrome (ARDS) and even death. The most common symptoms include headache, fatigue, fever, and cough.
The genome sequence of SARS-CoV-2 was determined, revealing 20 different proteins in the SARS-CoV-2 structure, aiding identification of infection mechanisms. Spike glycoprotein, a trimeric protein that attaches to the cell through angiotensin-converting enzyme 2 (ACE2), is one such protein.
Numerous nanostructures have been incorporated into therapeutic applications, including perovskites. These nanomaterials are notably used in medicine, such as photo-medicine and bioelectric implants.
Methodology
Researchers simulated the spike proteins that encounter a specific perovskite structure (SrTiO3, CaTiO3, BaTiO3) before ACE2. The effects of these perovskite structures on the deactivation of a spike protein were examined by employing coarse-grained MD simulations (CGMD).
The study employs CGMD in which small groups of atoms are assumed to be a single unit facilitating the investigation of larger timescales. This process is mainly used for polymeric chains materials. The researchers carried out molecular simulations on all atoms. CGMD and docking were conducted to test the interaction between the S protein of COVID-19 and the coarse PD of ACE2. Figure 1 shows the distorting in the structure of the spike protein.
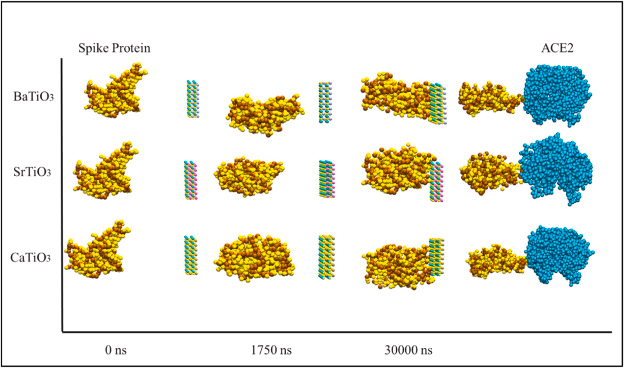
Figure 1. The process of encountering the spike protein with BaTiO3, CaTiO3, and SrTiO3 nanostructure in 0 ns, 1750 ns, and 3000 ns. The distorted spike protein then collides with the ACE2. Image Credit: Khedri, M., et al., 2021.
Simulation Details
MD simulations and data analyses were carried out with GROMACS software, and the energy was calculated by employing the molecular mechanics Poisson-Boltzmann surface area (MMPBSA) computational package.
The spike protein structure and ACE2 were accessed from RSCB, while the perovskite structures were designed via Avogadro software. This software was also used to develop the dimensions of the box and add the molecules and the solvent (the solvent is water to represent aqueous media.
Molecular Docking
The study used AutoDock_vina_1_1_2_linux_x86 for docking the simulation. The gasteiger charge and polar hydrogen were applied through the AutoDock Tool-1.5.6 to the SARS-CoV-2 RBD pdb and ACE2 pdb file, respectively.
Results
Energy
The nanostructure’s power to deform or distort the spike protein is shown by the intermolecular interaction energy. The energy between the nanostructures with spike protein has been calculated to restrain the S protein from penetrating through the ACE2 cell channel. Figure 2 depicts that both van der Waals (VdW) and the S protein’s electrostatic energy levels in the presence of BaTiO3, CaTiO3, and SrTiO3 nanostructures was decreased.
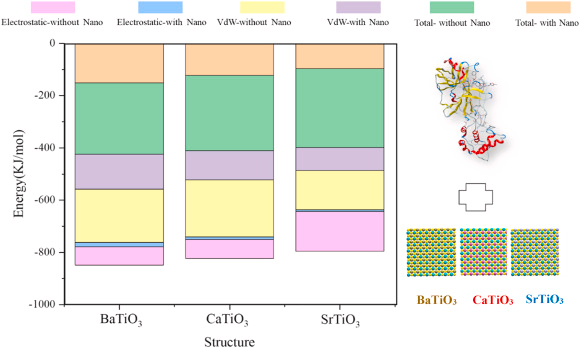
Figure 2. Electrostatic and van der Waals energy levels for Spike protein in direct contact with ACE2 and in the presence of BaTiO3, CaTiO3, and SrTiO3 nanostructures. Image Credit: Khedri, M., et al., 2021.
Upon exposure of the S protein to nano-perovskite structures, the absolute value of energy for re-docking of S protein and PD from ACE2 decreased significantly for all of the structures. The study details the average of the vital molecular analyzes (Table 1) carried out.
Table 1. The total energy, Rg, and average SASA of the spike protein encountered both ACE2 and perovskite nanostructures. Source: Khedri, M., et al., 2021.
| Total Energy (KJ/mol) |
| |
Energy with nanostructure |
Energy with ACE2 |
| BaTiO3 |
−150.613 |
−273.546 |
| CaTiO3 |
−121.647 |
−289.655 |
| SrTiO3 |
−95.974 |
−301.831 |
| Rg (ns) |
| BaTiO3 |
1.30072395 |
|
| CaTiO3 |
0.447702967 |
|
| SrTiO3 |
0.12459869 |
|
| Average of SASA (nm [2]) |
| BaTiO3 |
204.919495 |
|
| CaTiO3 |
206.55445 |
|
| SrTiO3 |
208.969104 |
|
Radius of Gyration
The radius of gyration details the compactness of the structure as a function of time. The results of the radius of gyration analyses in the presence of BaTiO3, CaTiO3, and SrTiO3 nanostructures to decrease the radius of gyration by contacting three nano-perovskite structures is shown in Figure 3.
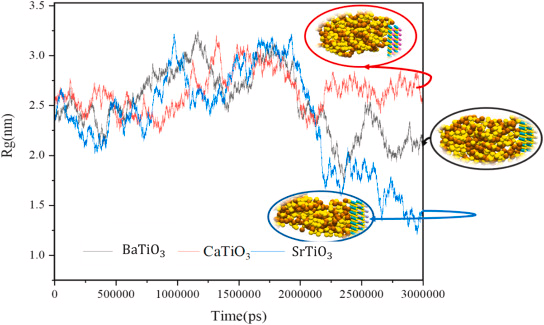
Figure 3. The radius of gyration for BaTiO3, CaTiO3, and SrTiO3 encountering spike protein. Image Credit: Khedri, M., et al., 2021.
RMSD and RMSF
RMSD demonstrates the flexibility of an atom to depart the S protein. The average distance between a certain spike protein and ACE2 in the presence and absence of perovskite nanostructure is shown in Figure 4a.
The average RMSD value between the S protein and ACE2 is higher than the S protein and ACE2 in the presence of perovskite nanostructure. A higher RMSD value indicates greater flexibility in the particle's movements due to thermal fluctuations.
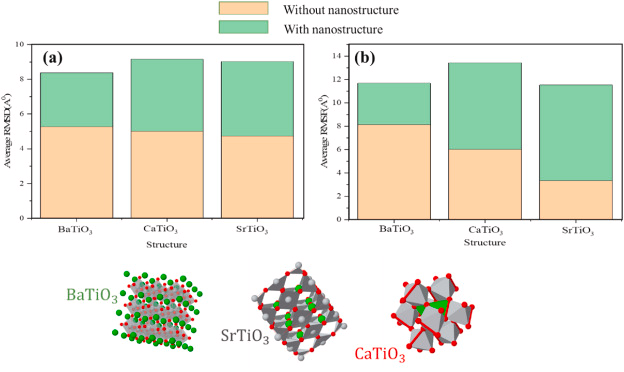
Figure 4. (a) RMSD and (b) RMSF for spike protein with and without BaTiO3, CaTiO3, and SrTiO3 nanostructures. Image Credit: Khedri, M., et al., 2021.
RMSF is the displacement in a single atom or a molecular structure of a reference atom or structure. Figure 4 b shows the RMSF value for the analyzed perovskite structures.
SASA
SASA defines the capacity of a material in an aqueous media to be surrounded by water molecules Figure 5 shows the SASA value of various nanostructures for adsorption of the spike protein.
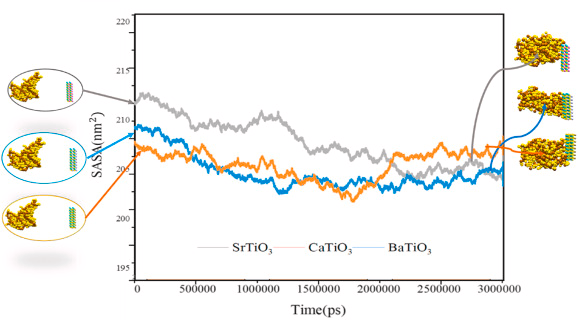
Figure 5. SASA of BaTiO3, CaTiO3, and SrTiO3 for adsorption of the spike protein. Image Credit: Khedri, M., et al., 2021.
Number of H-Bonds
Figure 6 depicts the number of intermolecular hydrogen bonds for the analyzed structures. During the absence of the perovskite nanostructures, hydrogen bonds are around 40, while Nano-perovskite decreases this value.
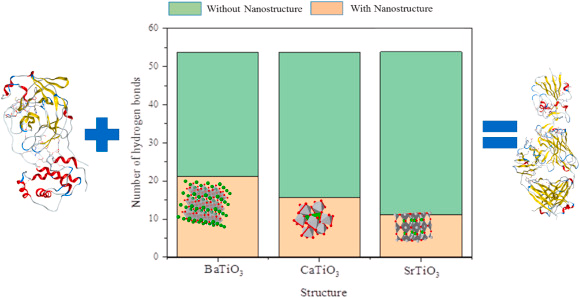
Figure 6. The number of hydrogen bonds between spike protein and ACE2 in the presence and absence of BaTiO3, CaTiO3, and SrTiO3 nanostructures. Image Credit: Khedri, M., et al., 2021.
Distribution of Secondary Structure (DSS)
The S protein's stability can be reduced by increasing the coil, bending, and turning, and decreasing the β-sheets and α-helices. Figure 7 shows that all the analyzed perovskites decrease the S protein’s stability.
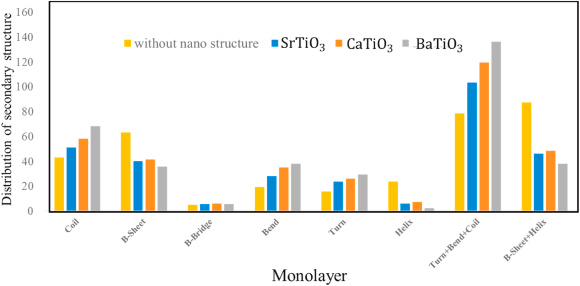
Figure 7. Distribution of secondary structures for S protein without and with BaTiO3, CaTiO3, and SrTiO3 nanostructures. Image Credit: Khedri, M., et al., 2021.
Discussion
Based on the energy diagram it can be deduced that SrTiO3 is better at reducing VdW and electrostatic energy levels when compared to the other nanostructures. SrTiO3 also tops as the best nano-perovskite structure based on the radius of gyration.
RMSD and RMSF analysis show that using a perovskite can limit the flexibility of the structure, which is undesirable. SrTiO3 is also the best for controlling the diffusion of S protein, along with reducing the number of hydrogen bonds more significantly than other nanostructures.
Conclusion
Examining the effects of various simulated nano-perovskite structures showed that nano-SrTiO3 can effectively prevent COVID-19 by distorting and deforming the S protein, thereby decreasing the possibility of penetration and viral propagation.
Continue reading: Cicada's Antimicrobial Nanotechnology Solution for COVID-Free Surfaces.
Journal Reference:
Khedri, M., Zandi, P. Ghasemy E., Nikzad, A., Maleki, R., Rezaei, N., (2021) In-silico study on perovskites application in capturing and distorting coronavirus. Informatics in Medicine Unlocked, 26, p. 100755. Available at: https://doi.org/10.1016/j.imu.2021.100755.
References and Further Reading
- Mouffouk, C., et al. (2021) Flavonols as potential antiviral drugs targeting SARS-CoV-2 proteases (3CLpro and PLpro), spike protein, RNA-dependent RNA polymerase (RdRp) and angiotensin-converting enzyme II receptor (ACE2). European Journal of Pharmacology, 891, p. 173759. doi.org/10.1016%2Fj.ejphar.2020.173759.
- Hamid, S., et al. (2020) Novel coronavirus disease (COVID-19): a pandemic (epidemiology, pathogenesis and potential therapeutics). New microbes and new infections, p. 100679. doi.org/10.1016/j.nmni.2020.100679.
- Gu, S X., et al. (2020) A pointer network based deep learning algorithm for unconstrained binary quadratic programming problem. Nature Reviews Cardiology, p. 1. doi.org/10.1038/s41569-020-00469-1.
- Yan, R., et al. (2020) Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science, 367(6485), p. 1444. doi.org/10.1126/science.abb2762.
- Mehalko, J., et al. (2021) Improved production of SARS-CoV-2 spike receptor-binding domain (RBD) for serology assays. Protein expression and purification, 179, p. 105802. doi.org/10.1016/j.pep.2020.105802.
- Khedri, M., et al., (2021).
- Perrella, F., et al. (2021) Interference of Polydatin/Resveratrol in the ACE2:Spike Recognition during COVID-19 Infection. A Focus on Their Potential Mechanism of Action through Computational and Biochemical Assays. Biomolecules, 11(7), p. 1048. doi.org/10.3390/biom11071048.
- Sitthiyotha, T & Chunsrivirot, S (2021) Computational design of SARS-CoV-2 peptide binders with better predicted binding affinities than human ACE2 receptor. Scientific Reports, 11(1), p. 1. doi.org/10.1038/s41598-021-94873-3.
- Kirchdoerfer, R N., et al. (2016) Pre-fusion structure of a human coronavirus spike protein. Nature, 531(7592), p. 118. doi.org/10.1038/nature17200.
- Rathod, S B., et al. (2020) Peptide modelling and screening against human ACE2 and spike glycoprotein RBD of SARS-CoV-2. In silico pharmacology, 8(1), p. 1. doi.org/10.1007/s40203-020-00055-w.
- Poland, G A., et al. (2020) SARS-CoV-2 vaccine development: current status. Mayo Clinic Proceedings. doi.org/10.1016/j.mayocp.2020.07.021.
- Chauhan G., et al. (2020) Nanotechnology for COVID-19: Therapeutics and Vaccine Research. ACS Nano, 14(7), p. 7760. doi.org/10.1021/acsnano.0c04006.
- Jonker, G & Van Santen, J (1950) Ferromagnetic compounds of manganese with perovskite structure. Physica, 16(3), p. 337. doi.org/10.1016/0031-8914(50)90033-4.
- Hodes, G (2013) Perovskite-Based Solar Cells. Science, 342(6156), p. 317. doi.org/10.1126/science.1245473.
- Xu, C., et al. (2021) Portable and wearable self-powered systems based on emerging energy harvesting technology. Microsystems & Nanoengineering, 7(1), p. 1. doi.org/10.1038/s41378-021-00248-z.
- Yu, D., et al. (2020) A Thermal Design of a 1 kW-Class Shell and Tube Methanol Steam Reforming System with Internal Evaporator. Processes, 8(11), p. 1509. doi.org/10.3390/pr8111509.
- Zhao, J., et al. (2020) Perovskite-filled membranes for flexible and large-area direct-conversion X-ray detector arrays. Nature Photonics, 14(10), p. 612. doi.org/10.1038/s41566-020-0678-x.
- Amati, G & Schilling, T (2020) Structural localization in the classical and quantum Fermi-Pasta-Ulam model. Chaos: An Interdisciplinary Journal of Nonlinear Science, 30(3), p. 033116. doi.org/10.1063/1.5130740.
- Helal, M A., et al. (2020) Molecular basis of the potential interaction of SARS-CoV-2 spike protein to CD147 in COVID-19 associated-lymphopenia. Journal of Biomolecular Structure and Dynamics, pp. 1–28. doi.org/10.1080/07391102.2020.1822208
- Basit, A., et al. (2020) Truncated human angiotensin converting enzyme 2; a potential inhibitor of SARS-CoV-2 spike glycoprotein and potent COVID-19 therapeutic agent. Journal of Biomolecular Structure and Dynamics, pp. 1–10. doi.org/10.1080/07391102.2020.1768150.
- Trott, O & Olson A J (2010) AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of computational chemistry, 31(2), pp. 455–461. doi.org/10.1002/jcc.21334.
- Bernshtein, V & Oref I (1998) Intermolecular energy transfer probabilities from trajectory calculations: A new approach. The Journal of chemical physics, 108(9), p. 3543. doi.org/10.1063/1.475750.
- Xiang, B., et al. (2020) Intermolecular vibrational energy transfer enabled by microcavity strong light–matter coupling. Science, 368(6491), p. 665. doi.org/10.1126/science.aba3544.
- Zhang, D & Lazim, R (2017) Application of conventional molecular dynamics simulation in evaluating the stability of apomyoglobin in urea solution. Scientific Reports, 7(1), p. 44651. doi.org/10.1038/srep44651.
- Kabsch, W & Sander C (1983) Dictionary of protein secondary structure: Pattern recognition of hydrogen‐bonded and geometrical features. Biopolymers: Original Research on Biomolecules, 22(12), p. 2577. doi.org/10.1002/bip.360221211.
- Qi, R., et al. (2014) Conformational distribution and α-helix to β-sheet transition of human amylin fragment dimer. Biomacromolecules, 15(1), p. 122. doi.org/10.1021/bm401406e.