Aug 10 2009
It's well known that the protein calmodulin specifically targets and steers the activities of hundreds of other proteins - mostly kinases - in our cells, thus playing a role in physiologically important processes ranging from gene transcription to nerve growth and muscle contraction But just how it distinguishes between target proteins is not well understood. Methods developed by biophysicists at the Technische Universitaet Muenchen (TUM) have enabled them to manipulate and observe calmodulin in action, on the single-molecule scale. In recent experiments, as they report in the early edition of PNAS, the Proceedings of the National Academy of Sciences, they compared the sequences of structural and kinetic changes involved in binding two different kinases. The results reveal new details of how calmodulin binds and regulates its target proteins.
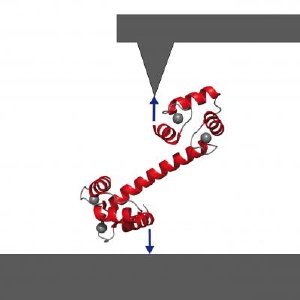 Biophysicists at the Tecdhnische Universitaet Muenchen use a specially designed atomic-force microscope to manipulate the signaling protein calmodulin and watch it do its work. Credit: M. Rief, TUM Dept. of Physics
Biophysicists at the Tecdhnische Universitaet Muenchen use a specially designed atomic-force microscope to manipulate the signaling protein calmodulin and watch it do its work. Credit: M. Rief, TUM Dept. of Physics
A so-called signaling protein and "calcium sensor," calmodulin gives start and stop signals for a great number of intracellular activities by binding and releasing other proteins. Calmodulin can bind up to four calcium ions, and the three-dimensional spatial structure of calmodulin varies with the number of calcium ions bound to it. This structure in turn helps to determine which amino acid chains - peptides and proteins - the calmodulin will bind.
Techniques such as X-ray structural analysis offer snapshots, at best, of steps in this intracellular work flow. But single-molecule atomic-force spectroscopy has opened a new window on such dynamic processes.
Professor Matthias Rief and colleagues at the Technische Universitaet Muenchen had previously shown that they could fix a single calmodulin molecule between a surface and the cantilever tip of a specially built atomic-force microscope, expose it to calcium ions in solution, induce peptide binding and unbinding, and measure changes in the molecule's mechanical properties as it did its work.
"What is special about our technique," Rief says, "is that we can work directly in aqueous solution. We can make our measurements in exactly the conditions under which the protein works in its natural environment. So we can directly observe how the calmodulin snatches the amino acid chain and folds itself, to hold its target fast." Measuring the force needed to bend the calmodulin molecule out of its stable condition at any given moment enables the researchers to compute the energies associated with binding both the calcium ions and the amino acid chains. And by following changes in the molecule's mechanical properties over time, they also can determine how long a protein fragment remains bound.
The results Rief and biophysicist Jan Philipp Junker report in the early edition of PNAS show that their approach also enables detailed comparative studies of binding sequences for different target proteins. The target sequences observed in these experiments are called skMLCK and CaMKK. Rief and Junker used mechanical force - actually pulling on complexes of calmodulin and the target peptides at rates of 1 nanometer per second or less - to slow down the processes to observable time scales and to clearly separate the individual unbinding steps.
"By applying mechanical force," Junker says, "we are able to dismantle the calmodulin-target peptide complex with surgical precision. Using conventional methods, this would be very difficult to do."
Among the detailed insights this approach made accessible are the hierarchy of folding and target binding, the sequence of unbinding events, and target-specific differences in terms of what is called cooperative binding.
Original paper: "Single-molecule force spectroscopy distinguishes target binding modes of calmodulin," by Jan Philipp Junker and Matthias Rief, published in the online Early Edition of PNAS, Proceedings of the National Academy of Sciences for the week of August 10, 2009.