Dec 22 2009
Cornell researchers are devising methods to detect the magnetic fields of individual electrons and atomic nuclei, which they hope to use to make a nanoscale version of magnetic resonance imaging.
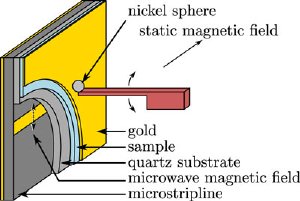 A schematic of John Marohn's scanned-probe electron spin resonance experiments, with the oscillating cantilever in the middle and the nickel tip that was attached by hand. Image: Eric Moore
A schematic of John Marohn's scanned-probe electron spin resonance experiments, with the oscillating cantilever in the middle and the nickel tip that was attached by hand. Image: Eric Moore
In biochemistry, shape is everything. Because of the shape of their binding sites, hormones can attach to cell membranes to send signals inside, and viruses can open up paths to invade. But understanding the structure of complex molecules, sometimes made of tens of thousands of atoms, is no easy task. Existing microscopes, even the world's best electron microscopes, can't resolve atomic details of organic material.
Researchers at Cornell are changing that: They are devising methods to detect the magnetic fields of individual electrons and atomic nuclei, which they hope to use to make 3-D images -- a nanoscale version of magnetic resonance imaging. Their new approach can detect a tiny force called electron spin to a sensitivity scale of about 400 electrons.
Their breakthrough is reported in the Dec. 14 online issue of Proceedings of the National Academy of Sciences.
"There is no more longstanding a problem than imaging the structure of membrane proteins," said John Marohn, associate professor of chemistry, who is leading the research and has been working on the problem for nearly a decade. "There is no general approach, and it is backbreaking work." Marohn belongs to a growing community of scientists doing single-molecule imaging, and in August Cornell hosted a conference on the topic.
At the heart of Marohn's research is the idea that electrons, protons and neutrons create a magnetic field, also called a magnetic moment. When placed next to a stronger magnetic field, all the little "magnets" -- electrons or neutrons and protons in the nuclei -- line up. Applying a radiofrequency field tuned to the electron or nucleus causes the magnetic moment to spin off axis like a wobbly top, a behavior called precession.
A medical MRI machine detects voltage induced in a coil by the precession of, typically, a thousand billion hydrogen atoms, which creates enough resolution for 3-D imaging of body parts. Marohn and colleagues are trying to make an instrument sensitive enough to feel the force of a single electron or nucleus in a protein.
In their paper, they describe their new approach, which involves detecting the spin of electrons in a molecule called a nitroxide -- rare because it contains a radical, or an unpaired electron. Their technique opens up the possibility of imaging, at sub-nanometer resolution, the locations of nitroxide "labels" attached to individual protein molecules.
By creating a sample of nitroxide molecules dissolved in a thin-film polymer and bringing the sample close to a 4-micron nickel magnet attached to a 350-nanometer silicon cantilever, they can detect the electron spin by measuring the frequency of the cantilever as it wobbles, like a diving board. The cantilever is similar to those used in scanning probe microscopy, a type of imaging that involves a cantilever scanning a surface and recording the probe-surface interactions.
To improve their frequency readouts and get more accurate measurements, the group must learn, among others things, how to make their magnets smaller, Marohn said.
"The dream is to take that signal, run it through some sort of complicated algorithm, and get a picture," Marohn said.