One process used to produce nanoscopic structures like ever-smaller integrated circuits, biosensors, and gene chips is known as dip-pen nanolithography, in which the nanotip of an atomic force microscope is used to "write" a pattern directly on a substrate. In the journal Angewandte Chemie, a Korean research team led by Jung-Hyurk Lim at Chungju National University in Chungju have now introduced a refined nanotip for this technique.
With their "nanoquill", it is possible to produce complex nanopatterns from large biomolecules - such as complete virus particles - rapidly, precisely, and flexibly.
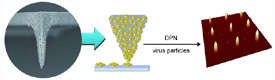
Atomic force microscopy, originally designed for the determination of the nanoscopic structures of surfaces, has since been very successfully put to another use: In dip-pen nanolithography, the nanotip is dipped like a quill into an "ink well" and the molecules are then deposited like ink onto a suitable substrate to form complex nanopatterns. Critical to this process is a tiny water meniscus that forms between the surface to be written on and the nanotip; the meniscus provides a pathway by which the molecules in the ink - DNA, peptides, or proteins - can move to the surface. However, larger molecules cannot diffuse through the meniscus and cannot be deposited on the surface. Thanks to a novel nanotip, the Korean scientists have now overcome this limitation.
The new tip is made of silicon dioxide that has been coated with a well-characterized biocompatible polymer. This forms a nanoporous polymer network with pore diameters between 50 and several hundred nanometers.
When this tip is dipped into a solution containing biomolecules, the polymer absorbs the liquid and swells into a gel. When the loaded "nanoquill" comes into contact with an amine-coated substrate, the biomolecules diffuse out of the gel onto the surface. Because diffusion from the gel onto the surface encounters less resistance than diffusion through a water meniscus, it is possible to deposit much larger biomolecules than in the conventional method.
As a demonstration, the researchers selected virus particles bound to a fluorescence dye as their ink. They were able to use this to produce patterns with more than 1000 individual nanodots without having to refill the quill. Unlike the conventional technique, increasing contact time between the surface and the tip of the quill increases the number of individual viruses within the dot, but not its diameter. However, the researchers were able to generate dots of various sizes (400, 200, and 80 nm) by varying the diameter of the tip. This variation can be quite easily controlled by the duration of the polymerization reaction.