Feb 28 2017
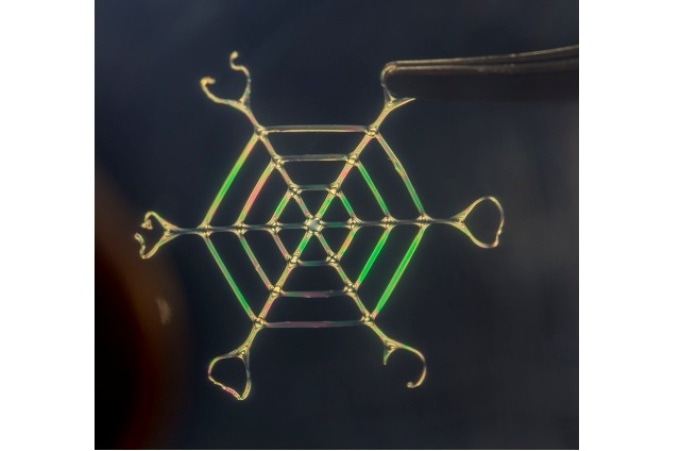 A bio-inspired web made of silk nano fibers, 2 to 3 cm in width. CREDIT: Silk Lab Tufts University.
A bio-inspired web made of silk nano fibers, 2 to 3 cm in width. CREDIT: Silk Lab Tufts University.
A research team from the School of Engineering at Tufts University has invented an innovative bioinspired method to transform silk protein into robust and ultralight complex materials that can be easily programmed at the macro-, micro-, and nano-scales.
A web of silk nano fibers was one among numerous structures developed by the researchers, where the silk nano fiber web can endure a load of up to 4000 times its weight. The study has been published online in the 27 February issue of the Nature Nanotechnology journal.
It is a known fact that structural proteins are nature’s building blocks, which have the ability to form materials that provide structure, stiffness, and function in biological systems.
One of the main challenges in developing similar synthetic materials is the hierarchical structure of natural materials, which presents peculiar properties at both the macro and molecular levels. Efforts put by researchers in emulating this structure results in the finding that control at one scale interferes with control at other scales.
The research team from the Tufts University integrated the bottom-up, self-assembly property of the natural materials with the top-down, directed assembly to concurrently control micro-mechanical constraints, geometry at all scales, and solvent-removal dynamics which regulate the biomaterial properties.
We generated controllable, multi-scale materials that could be readily engineered with dopant agents. While silk is our main focus, we believe this approach is applicable to other biomaterials and composites and synthetic hydrogels.
Fiorenzo Omenetto, PhD, Tufts University
Omenetto is the Frank C. Doble Professor in the Department of Biomedical Engineering, and the corresponding author of the study. He is a part of the Department of Electrical and Computer Engineering and to the Department of Physics within the School of Arts and Sciences.
The new method was employed to pattern centimeter-scale silicone molds that include micro-scale features with thickness less than or equal to human hair. The molds were injected with an aqueous fibroin protein gel extracted from silkworm cocoons and were mechanically stressed by performing contraction of the gel using ethanol and water and/or by physically deforming the entire mold.
Upon drying, the structure of the silk protein gets naturally transformed into a highly robust beta-sheet crystal. The mechanical properties and final shape of the material were accurately engineered by regulating the micro-scale pattern of the mold, the gel contraction, the deformation of the mold, and silk dehydration.
The final result of our process is a stable architecture of aligned nanofibers, similar to natural silk but offering us the opportunity to engineer functionality into the material.
Peter Tseng, PhD, Postdoctoral Scholar in Omenetto’s Silk Lab at the School of Engineering of the Tufts University
In the case of specific experiments, the silk gel was doped with gold nanoparticles. The research team performed this step as gold nanoparticles have the ability of transporting heat upon exposure to light.
Tseng indicated that spiders spin webs that are not porous but structurally dense. “In contrast, our web structure is aerated, porous and ultra-light while also robust to human touch, which may enable every-day applications in the future,” he added. A web with a diameter of 2 to 3 cm and weighing nearly 2.5 mg had the ability to support a weight of 11 g.
Other paper authors included Bradley Napier, Tufts doctoral student in the Silk Lab; Siwei Zhao, Ph.D., post-doctoral associate in the Silk Lab; Alexander N. Mitropoulos, Ph.D., former Tufts doctoral student in biomedical engineering, now at the United States Military Academy at West Point; Matthew B. Applegate, Ph.D., former Tufts doctoral student in biomedical engineering, now at Boston University; Benedetto Marelli, Ph.D., former post-doctoral associate in the Silk Lab, now at MIT; and David L. Kaplan, Ph.D., Stern Family Professor of Engineering. Kaplan has additional Tufts faculty appointments in the Department of Chemical and Biological Engineering, the Department of Chemistry in the School of Arts and Sciences, the School of Medicine and the School of Dental Medicine.
The Office of Naval Research partially supported the research under award N000141310596. The National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health supported Peter Tseng under the Kirschstein National Research Service Awards fellowship number F32EB021159-02. The research was also supported by the Air Force Office of Scientific Research.