Determining the exact location of an object within a tissue sample via surface-enhanced spatially offset Raman spectroscopy (SESORS) imaging is an emerging research interest to confirm its implementation for in vivo diagnostics in clinical settings.
Study: Tomographic Imaging and Localization of Nanoparticles in Tissue Using Surface-Enhanced Spatially Offset Raman Spectroscopy. Image Credit: Nomad_Soul/Shutterstock.com
In an article recently published in the journal ACS Applied Materials and Interfaces, researchers utilized spatially offset Raman spectroscopy (SORS) to investigate multiple experimental factors relevant to the optical setup in imaging.
The effects of spatial offset magnitude and geometry were reported in determining the location of nanoparticles mixed with silica powder. The nanoparticles were used as imaging targets through tissue which helped interpret SESORS images to assign the exact location of nanoparticles in a two-dimensional (2D) x, y-imaging plane located at a depth. Moreover, the linear offset-induced image drag effect highlighted the spatial distortion in SESORS images caused due to the direction and magnitude of the linear offset.
This spatial distortion in images necessitated neutralizing the asymmetric effects via annular SORS collection geometry during imaging.In addition, ratiometric SESORS imaging helped locate the buried targets in a three-dimensional (3D) model.
These methods aided in locating the surface-enhanced Raman scattering (SERS)-active inclusions (nanoparticles) in 3D models. With the help of spatial offset, probed depth, and ratiometric analysis of Raman intensities of the nanoparticle and tissue, the SESORS technique could image and discriminate between two different nanoparticles buried at varying depths within a 3D model.
Raman Techniques Towards Bioimaging
SESORS is an analytical technique that combines the SERS Raman signal enhancement effect with the through-barrier detection capability of SORS. This combination of SERS and SORS to produce SESORS enhances the transformational potential of Raman-based techniques for in vivo detection of various diseases at different stages.
SORS spectrometers use a spatial offset with a magnitude denoted as dx. During measurements, this spatial offset on the obscuring barrier facilitates the collection of Raman photons scattered by subsurface analytes. The multilayered biological samples, for example, mammalian tissue analogs, could be excited when incorporated with SERS active nanoparticles and subsurface-scattered Raman photons.
SERS is a plasmonic effect where the molecules adsorbed on a rough metal surface can result in high Raman scattering intensities. Various nanoparticles are used for SERS applications and the SERS effect increases Raman signal intensity by up to 1014 to 1015-fold. Gold nanoparticles with the SERS effect are suitable for biomedical applications.
The SESORS-based imaging technique is used to detect various disease states in biological tissues, where a biomolecular target interacts with biofunctionalized nanoparticles (incorporated with SERS activity) and produces a specific molecular fingerprint spectrum. Tracking the intensity of the SERS band of concerned nanoparticles helps in the bioimaging of biological samples that ascertains the location of nanoparticles in the 2D imaging plane at a depth.
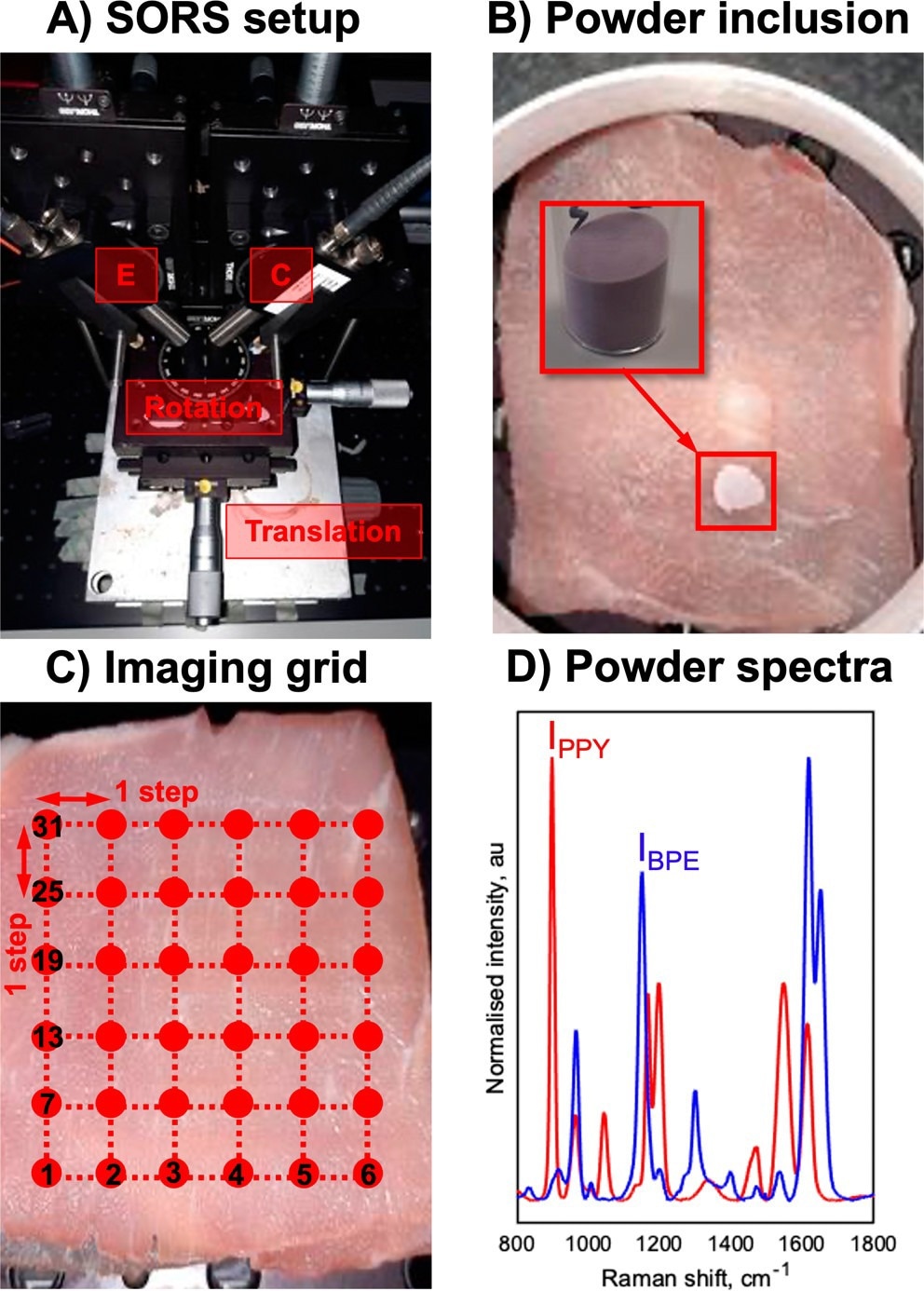
(A) SESORS experiments were conducted on an in-house-built SORS system with a point-based collection geometry, i.e., with individual excitation (E) and collection (C) probes, developed by Asiala et al. based on the design of Shand and co-workers. (21,23) (B) Templates for SESORS experiments were prepared by mixing the silica powder with silica-coated SERS-active NPs. The NP powders were then added to a section of the porcine tissue and then covered with further tissue sections. (C) Hypothetical SESORS imaging regime illustrated using a square grid, where each vertex on the grid represents a pixel where the tissue surface is excited by the laser. In the regime illustrated, the grid represents a 36-pixel (6 × 6) image. (D) Truncated, baselined, and normalized SERS spectra of PPY (red) and BPE (blue) powder-based imaging templates developed for this study. Spectra were collected for 0.1 s using a 785 nm, 400 mW laser. © Berry, M. E., McCabe, S. M., Sloan-Dennison, S., Laing, S., Shand, N. C., Graham, D., Faulds, K. (2022).
Localization of Nanoparticles with SERS activity in Tissue Using SESORS
In the present study, a novel SESORS imaging template was utilized through outline and tissue experimental techniques to demonstrate the effect of spatial offset magnitude and geometry in locating the nanoparticles with SERS activity. This technique allowed the precise interpretation of the location of nanoparticles with SERS activity in a 2D plane with the help of SESORS images.
The magnitude and direction of the linear offset vector cause a spatial distortion in SESORS images, represented as linear offset-induced image drag. Consequently, this necessitated the imaging with radial SORS collection geometry, thereby neutralizing the effects of asymmetric image artifacts to avoid data misinterpretation and inaccurate identification of inclusion’s location.
Simultaneously, introducing the ratiometric SESORS imaging concept helped locate buried nanoparticle inclusions in 3D structures. This novel technique differed from conventional SESORS imaging in terms of pixels quantification, wherein the subsurface nanoparticle SERS band helped pixel quantification.
The relative contribution of the SERS band of nanoparticles through tissue spectrum (intensity through nanoparticles (INP) / intensity through tissue (ITis)) could quantify each pixel’s value in building the image. Furthermore, the robustness of this method was tested by utilizing the correlation between spatial offset’s magnitude, the probed depth, and INP/ITis ratiometric analysis for imaging and spatially discriminating between two different nanoparticles buried at varying depths in a 3D structure.
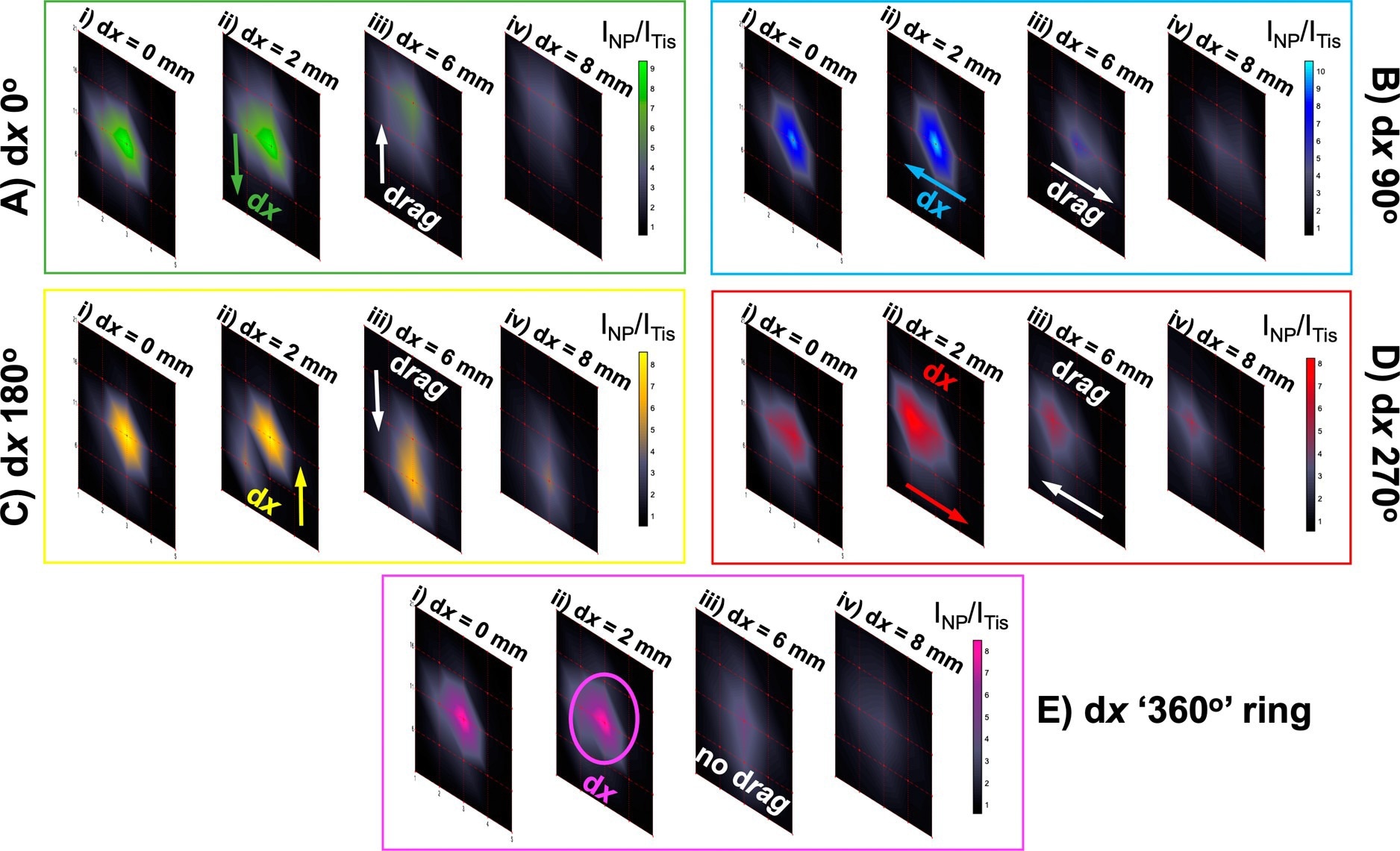
Probing through tissue in three dimensions using ratiometric SESORS imaging and a variable ring-collection offset. (A) Ratiometric images were collected of the PPY powder buried beneath 6 mm of tissue across a 20 × 20 mm grid in 5 mm step sizes with linear offset magnitudes of (i) 0, (ii) 2, (iii) 6, and (iv) 8 mm. This was repeated three times with the sample being rotated through 360° in 90° steps to mimic the rotation of the linear offset vector, and the resulting vectors were termed with respect to the orientation of the vector in the initial set of images (dx “0°”) as (B) dx “90°”, (C) dx “180°”, and (D) dx “270°”. (E) Ratiometric images averaged across the four linear offset vectors to mimic a ring-collection offset at (i) 0, (ii) 2, (iii) 6, and (iv) 8 mm. The color bars across each set of offset images were scaled for clarity to indicate the offset that maximized the intensity ratio, INP/ITis. Spectra were collected for 1 s using a laser power of 400 mW. © Berry, M. E., McCabe, S. M., Sloan-Dennison, S., Laing, S., Shand, N. C., Graham, D., Faulds, K. (2022).
Conclusion
In summary, the accurate interpretation of SESORS images helped locate the precise position of targeted nanoparticles with SERS activity in a 3D model, which could be applicable in disease state detection. Investigating multiple factors relevant to optical settings for tissue imaging using a SORS system determined the magnitude and geometry of spatial offset in locating the nanoparticles with SERS activity through the tissue.
Furthermore, a linear offset-induced image drag effect was presented, highlighting the SESORS image’s spatial distortion caused by the direction and magnitude of the linear offset vector. Consequently, an annular SORS collection geometry during imaging removes the asymmetric drag effects in images to accurately locate the nanoparticles with the SERS effect.
Reference
Berry, M. E., McCabe, S. M., Sloan-Dennison, S., Laing, S., Shand, N. C., Graham, D., Faulds, K. (2022). Tomographic Imaging and Localization of Nanoparticles in Tissue Using Surface-Enhanced Spatially Offset Raman Spectroscopy. ACS Applied Materials & Interfaces. https://pubs.acs.org/doi/10.1021/acsami.2c05611
Disclaimer: The views expressed here are those of the author expressed in their private capacity and do not necessarily represent the views of AZoM.com Limited T/A AZoNetwork the owner and operator of this website. This disclaimer forms part of the Terms and conditions of use of this website.