The most powerful technique for establishing the three-dimensional structures of biological macromolecules is Macromolecular crystallography (MX). It usually gives the most reliable structures and attains the highest-resolution information, and does not suffer from limitations on sample size as long as crystals are available.
Most diffraction data are gathered at synchrotron beamlines with fast detectors, high photon flux, powerful automation, highly experienced staff and expert processing pipelines.
Today, almost two thirds of all structures are established using PILATUS and EIGER HPC detectors, as seen in Figure 1. Their beneficial properties, like maintenance-free operation and noise-free readout, make these detectors ideal for academic laboratories too.
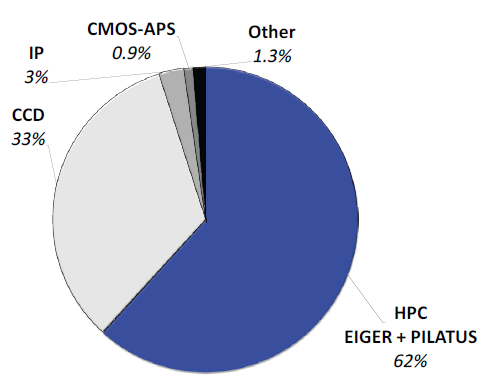
Figure 1. Contribution of detectors to PDB entries released in 2019. Nearly two thirds of all structures were solved with data collected with PILATUS and EIGER HPC detectors. Legacy detectors based on CCDs and image plates (IP) make up most of the rest. Competing technology such as CMOS-APS plays no role. Image Credit: DECTRIS Ltd.
In many instances, laboratory diffractometers which are equipped with PILATUS and EIGER detectors are useful for a lot more than optimizing crystallization and cryo-cooling conditions before data collection at a synchrotron.
This is especially true for structural enzymology work where data on mutants is used to create new mutations to probe the active site and for drug design projects which need a fast turn-around in response to newly synthesized compounds. Below, some recent results in these two fields of structural biology are discussed.
Drug Development
Rational Design of Antisickling Agents
Sickle cell disease is a hereditary blood disorder associated with episodes of pain, infections, anemia and delayed growth and development. The red blood cells are deformed into the shape of a sickle in patients with this disease.
It is caused by mutations in the gene coding for beta-globin, which is one of the two components of the hemoglobin tetramer. At Virginia Commonwealth University, the laboratory of Prof. Martin Safo studies the structure-function relationship of hemoglobin with the aim of developing antisickling agents.
Medicinal chemists design ligands that are tested for their pharmacology in vivo and in vitro, in tight collaboration with crystallographers. Structures of hemoglobin cocrystallized with promising candidates are solved quickly from diffraction data gathered with Prof. Safo‘s EIGER R 4M detector, as seen in Figure 2.
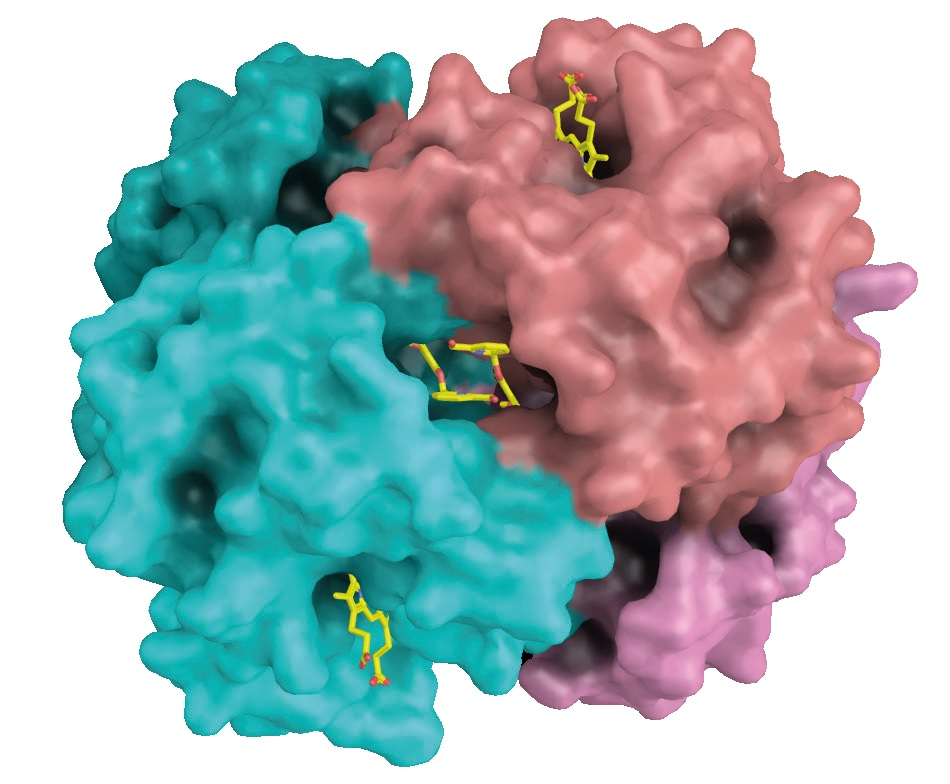
Figure 2. The antisickling agent SAJ-009 binds at the interface between two alpha-globin chains. The alpha-globins are shown in cyan and salmon. Heme groups bind to pockets in the alpha-globin chains (top right and bottom left). Image Credit: DECTRIS Ltd.
The ability to react quickly to new information is one of the key benefits of having structural biology and medicinal chemistry in close proximity. Prof. Safo’s group can solve structures in a couple of days instead of waiting for synchrotron time.
Structural Biology in the Pharmaceutical Industry
For pharmaceutical research, development and quality control during production, powerful laboratory diffractometers are crucial. They enable all aspects of the experiment to be under total control and for decisions to be made fast.
Among others, Novo Nordisk in Denmark, Astex Pharmaceutical in the UK, and Array Biopharma in the US, are all productive with state-of-the-art diffractometers with EIGER R 1M or PILATUS3 R 1M detectors.
Fighting Trypanosoma Diseases
In developing countries, Chagas’ disease, sleeping sickness, and leishmaniasis pose serious health problems. All of these diseases are caused by trypanosomes, protozoans characterized by a unique trypanothione redox system.
At the University of Toronto, Prof. Emil Pai’s laboratory studies trypanothione reductase inhibitors, which is an attractive class of ligands inactive against organisms with conventional glutathione redox systems.
Prof. Pai’s group developed the strongest competitive trypanothione reductase inhibitor reported to date, starting from a previously reported structure. The new inhibitor binds into a large active site, which is occupied simultaneously by a buffer molecule retained from protein purification, as seen in Figure 3.
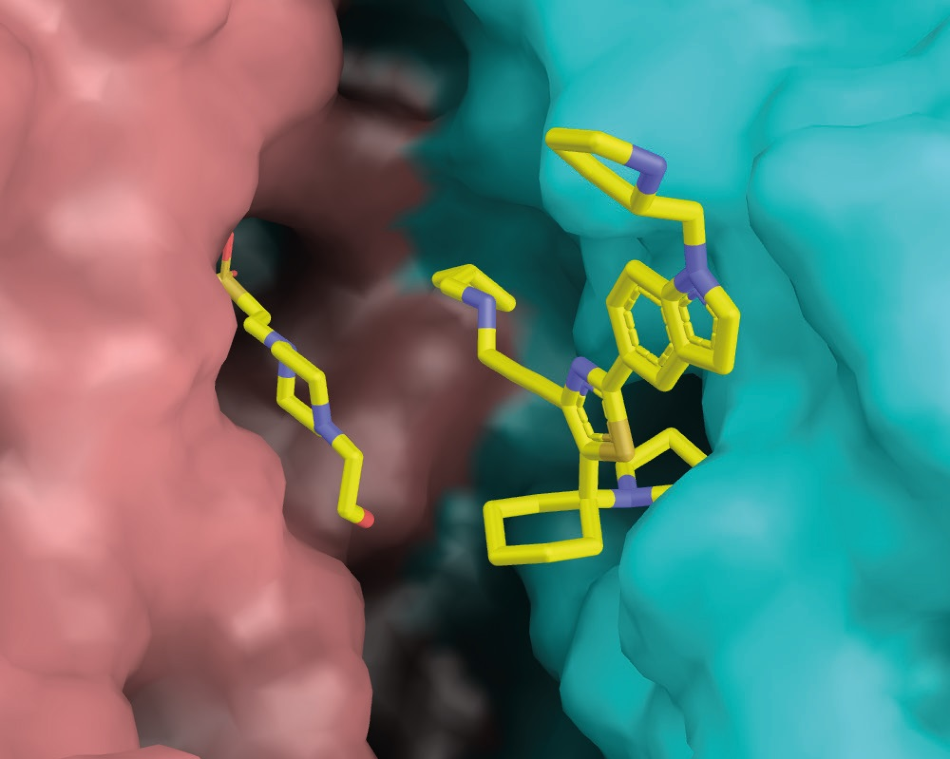
Figure 3. Ligand (+)-2 binds at the dimer interface of trypanothione reductase.(TR). The two TR protomers are shown in salmon and cyan, respectively. On the left of the binding pocket, an accidentally bound HEPES molecule can be seen. Image Credit: DECTRIS Ltd.
Data collection was straightforward despite the large unit cell of the crystal (118 x 118 x 225 Å3) due to the single-pixel point spread and the small pixels of EIGER R 1M. An attractive goal for further research is combining the two ligands into one even more tightly binding lead compound.
Structural Enzymology
Catalytic Mechanism of L-asparaginases
L-asparaginases are enzymes that catalyze the hydrolysis of the amino acid asparagine to aspartate. They are crucial for the treatment of selected lymphomas and leukemias.
At the National Cancer Institute in Frederick, Dr. Alexander Wlodawer’s laboratory studies the catalytic mechanism of L-asparaginases. Understanding of the mechanism is dependent on the correct interpretation of all of the components of the active site.
Six crystal structures established in Dr. Wlodawer’s laboratory from data gathered on EIGER R 4M detectors exhibit how the earlier structures were misinterpreted as containing native ligands when the electron density seen can be better explained with an extraneous ligand that bound to the enzyme during crystallization, as seen in Figure 4.
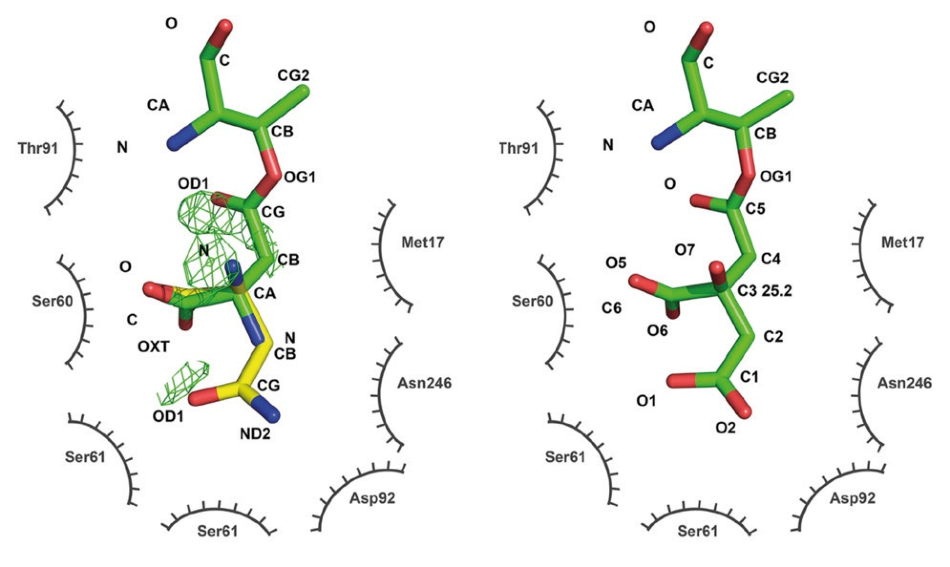
Figure 4. Active site of L-asparaginase. An older structure (PDB code 2HIM, left) was interpreted as showing aspartate covalently bound to threonine 14 (green, catalytic intermediate) with a substrate asparagine overlaid (yellow). Difference density indicates problems with the model. The new structure (PDB code 6NXC, right) reinterprets the ligand density as a covalently bound citrate from the crystallization buffer. No difference density remains after refinement. Figure modified from Lubkowski et al. 2019 (Reference 2, CC BY 4.0). Image Credit: DECTRIS Ltd.
Catalytic Mechanism of Nitrite Reductases
The group of Prof. Samar Hasnain and Dr. Svetlana Antonyuk at the University of Liverpool, as seen in Figure 5, have studied the chemistry of nitrite reductases for a long time. These proton-coupled redox enzymes are susceptible to radiation damage as they contain multiple metal centers.
Recent lab work combines experiments at multiple radiation sources in order to elucidate the catalytic pathway of nitrite reductases. Experiments at a neutron source to show the protonation states of the catalytic residues, at an X-ray free-electron laser to show the undamaged active site, and at the laboratory diffractometer to determine the resting states at different pH values were of particular importance.
Conclusion
Detector upgrades have helped academic and industrial laboratories answer biological questions faster and to use their synchrotron time more wisely.
An ideal tool for studying ligand binding in drug discovery projects and establishing the influence of individual side chains on the chemistry of an enzyme quickly is a diffractometer with a state-of-the-art detector. Structures can be solved by native SAD and the effects of pH and temperature changes on a structure can be judged.
A modern diffractometer is also ideal for the training of the next generation of crystallographers. A laboratory diffractometer can be much more than a screening tool for finding the best crystallization and cryo-protection conditions.
Publication-quality structures can be acquired in the time it takes to send crystals to a synchrotron. In the coming years, when many synchrotrons undergo upgrade projects with extended shutdown periods, this will be particularly important.
Speed up crystallography by upgrading an existing source with a new detector or by getting a new diffractometer. One possible partner is marXperts, who upgraded the diffractometer in Liverpool with an EIGER R 4M detector, as seen in Figure 5.

Figure 5. The group of Dr. Antonyuk and Prof. Hasnain in front of their upgraded diffractometer. The EIGER R 4M is mounted on a mardtb (marXperts, Germany). Image Credit: DETCRIS Ltd.
References and Published Structures
- T. M. Deshpande et al., Acta Cryst D. 74, 956-964 (2018), doi:10.1107/S2059798318009919. 6DI4 (EIGER R 4M).
- A. Nakagawa et al., Molecular Pharmaceutics. 15, 1954- 1963 (2018), doi:10.1021/acs.molpharmaceut.8b00108. 6BWP, 6BWU (EIGER R 4M).
- P. P. Pagare et al., Bioorganic & Medicinal Chemistry. 26, 2530-2538 (2018), doi:10.1016/j.bmc.2018.04.015. 6BNR (EIGER R 4M).
- J. B. Fell et al., ACS Med. Chem. Lett. 9, 1230-1234 (2018), doi:10.1021/acsmedchemlett.8b00382. 6N2K, 6N2J (PILATUS3 R 1M).
- T. D. Heightman et al., J. Med. Chem. 62, 4683-4702 (2019), doi:10.1021/acs.jmedchem.9b00279. 6QMC, 6QMK (EIGER R 1M).
- A. M. Kidger et al., Molecular Cancer Therapeutics, online first (2019), doi:10.1158/1535-7163.MCT-19-0505. 6RQ4 (EIGER R 1M).
- A. Oddo et al., Biochemistry. 57, 4148-4154 (2018), doi:10.1021/acs.biochem.8b00105. 5OTX, 5OTW, 5OTV, 5OTU (PILATUS3 R 1M).
- R. De Gasparo et al., Chemistry-A European Journal. 25, 11416-11421 (2019), doi:10.1002/chem.201901664. 6OEZ (EIGER R 1M).
- J. Lubkowski et al., Protein Science. 28, 1850-1864 (2019), doi:10.1002/pro.3709. 6PA2, 6PA4, 6PA5, 6PA6, 6PA8, 6PA9, 6PAA, 6PAB, 6PAC, 6PAE (EIGER R 4M).
- J. Lubkowski et al., Scientific Reports. 9 (2019), doi:10.1038/s41598-019-46432-0. 6NX6, 6NX7, 6NX8, 6NX9, 6NXA, 6NXB (EIGER R 4M).
- T. P. Halsted et al., IUCrJ. 6, 761-772 (2019), doi:10.1107/S2052252519008285. 6GTL, 6GTN, 6GTI, 6GTK (EIGER R 4M).
- T. M. Hedison et al., ACS Catalysis. 9, 6087-6099 (2019), doi:10.1021/acscatal.9b01266. 6QPU (EIGER R 4M).
Acknowledgments
- Produced from materials originally authored by Dr. Andreas Förster from DECTRIS Ltd.

This information has been sourced, reviewed and adapted from materials provided by DECTRIS Ltd.
For more information on this source, please visit DECTRIS Ltd.