Jul 2 2009
Decades of research have failed to conclusively crack the mystery of how amyloid-associated diseases like Alzheimer's do so much damage, but new findings by researchers at RIKEN suggest that part of the answer may lie in the structural rearrangements observed in plaque-forming proteins.
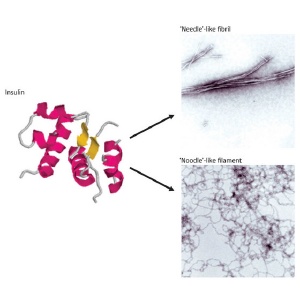 Structural differences of the insulin protein. Under different chemical conditions, the normally helical insulin protein rearranges into amyloid plaques composed of stiff ‘needle-like’ fibers or floppy ‘noodle-like’ filaments, with different physiological effects.
Structural differences of the insulin protein. Under different chemical conditions, the normally helical insulin protein rearranges into amyloid plaques composed of stiff ‘needle-like’ fibers or floppy ‘noodle-like’ filaments, with different physiological effects.
Although there is a 'preferred' structure for any particular protein, certain conditions can cause proteins to rearrange into new conformations that may dramatically alter their activity. For instance, the pathology of many serious diseases—including Parkinson's and Alzheimer's diseases—has been linked with formation of specific proteins into dense, clumps of fibrils known as amyloid plaques.
The hormone insulin, normally composed of two helical subunits, undergoes dramatic rearrangements under acidic conditions that cause it to form toxic amyloid fibrils. Although this transition is well known, Tamotsu Zako and his colleagues at the RIKEN Advanced Science Institute in Wako were recently taken aback to find that bovine insulin undergoes a different type of rearrangement when prepared in the presence of the reducing agent tris(2-carboxyethyl)phosphine, forming spidery filaments rather than stiff fibrils*.
In its fibrillar state, insulin loses its helical characteristics and assumes a structure known as a ß-sheet, a two-dimensional strip composed of multiple linear protein strands. On closer analysis, Zako's team noted that insulin subunits within filaments also assume a ß-sheet conformation. However, they noted a subtle difference: in filaments, these sheets are antiparallel rather than parallel, consisting of aligned polypeptide chains that zigzag back and forth rather than running in the same direction.
There was a more important difference, however. When cultured cells were treated with increasing doses of fibrillar insulin, they died in greater numbers, but no such toxicity was observed from filamentous insulin. "It was very unexpected and surprising that the cell toxicity of 'noodle-like' insulin was much different from 'needle-like' fibrils," Zako says.
This suggests that the secret of amyloid toxicity may lie in the structure of the protein aggregates, perhaps resulting from direct physical damage to cell membranes by fibrils. Zako's team is now attempting to resolve the basis for these differences in toxicity through structural analysis, but is also examining characteristics of another protein with similar behavior to see whether their findings may have broader implications for other amyloid-forming proteins linked with human pathologies, such as Alzheimer's-associated amyloid-ß protein. "We want to know whether other filamentous amyloids are also non-toxic," he says. "If so, it will become possible to generalize our findings."
*Zako, T., Sakono, M., Hashimoto, N., Ihara, M. & Maeda, M. Bovine insulin filaments induced by reducing disulfide bonds show a different morphology, secondary structure, and cell toxicity from intact insulin amyloid fibrils. Biophysical Journal 96, 3331-3340 (2009).