CRAIC Technologies is the worlds leading developer of UV-visible-NIR range scientific instruments for microanalysis. These include the QDI series UV-visible-NIR microspectrophotometer instruments designed to help you non-destructively measure the optical properties of microscopic samples. CRAIC's UVM series microscopes cover the UV, visible and NIR range and help you analyze with sub-micron resolutions far beyond the visible range. CRAIC Technologies also has the CTR series Raman microspectrometer for non-destructive analysis of microscopic samples. And don't forget that CRAIC proudly backs our microspectrometer and microscope products with unmatched service and support.

UV-Visible Microspectroscopy of Protein Crystals
UV-visible range microspectroscopy is an important technique for analysis of individual protein crystals. Currently, the protein crystals are used for structural biology and drug design, controlled drug delivery and bioseparations. As such, UV microspectroscopy is used for both the study of individual crystals optical properties as well as for quality control of crystals as they are grown.
Below is an example of the UV-visible microspectra of individual protein crystals. Additionally, the fluorescence emission spectra were also measured of the same crystals. All measurements were made with a QDI 2010™ UV-visible-NIR range microspectrophotometer. The crystals were grown in quartz capillary tubes and the spectra were taken of the crystals while still in the capillary tubes.
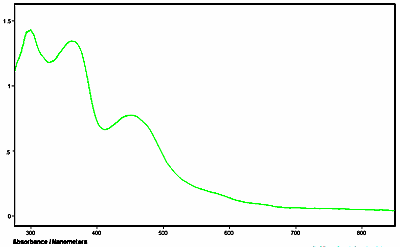
Figure 1. Absorbance spectrum of protein crystal taken with a QDI 2010™ UV-visible microspectrophotometer
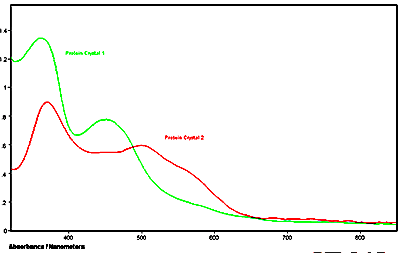
Figure 2. Absorbance spectra of two different proteins in crystalline form acquired on a QDI 2010™ MSP
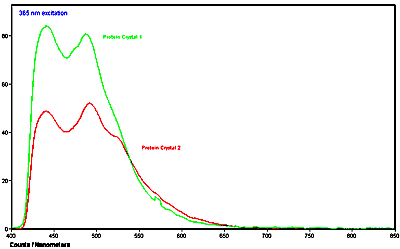
Figure 3. Fluorescence emission spectra of two different proteins in crystalline form acquired with 365 nm excitation taken with a QDI 2010™ microspectrophotometer
Primary author: Dr. Paul Martin
Source: UV-visible Microspectroscopy of Protein Crystals by CRAIC Technologies.

This information has been sourced, reviewed and adapted from materials provided by CRAIC Technologies.
For more information on this source, please visit CRAIC Technologies.