Aug 6 2009
Metal nanoparticles have radically different electronic, optical and magnetic properties from their larger states, which makes them useful as materials in new, ultra-small devices such as biological sensors. Constructing such devices, however, is difficult because, unlike atoms, nanoparticles lack directional bonds that allow them to be arranged precisely.
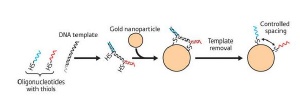 Combining reactive oligonucleotides with a DNA template allows controlled positioning of the nucleic acids on the surface of a gold nanoparticle.
Combining reactive oligonucleotides with a DNA template allows controlled positioning of the nucleic acids on the surface of a gold nanoparticle.
One strategy to overcome this limitation is to attach oligonucleotides—single strands of molecules that constitute DNA—to nanoparticle surfaces, and then, through Watson–Crick base pairing of the nucleic acids, join the nanoparticles together. However, manipulating the number and positions of oligonucleotides on the nanoparticles has been impossible.
Now, Kenji Suzuki, Kazuo Hosokawa and Mizuo Maeda from the RIKEN Advanced Science Institute in Wako have developed a method to immobilize oligonucleotides on gold nanoparticle surfaces with precise control over their number and geometric arrangement*. Because this procedure can be used for nanoparticles other than gold, it should initiate improved techniques for spontaneous assembly of small materials into complex structures—so-called ‘bottom–up’ nanotechnologies.
In their proof-of-principle experiment, Suzuki and colleagues combined two oligonucleotides containing reactive thiol (sulfur-hydrogen) groups with a third, non-thiolated oligonucleotide template to create a DNA nanostructure (Fig. 1). This DNA template was then reacted with a gold nanoparticle, forming a complex through the active thiol groups. Finally, the DNA template was separated from the complex, leaving two free oligonucleotide strands on the gold nanoparticle.
Transmission electron microscopy imaging confirmed the success of the DNA template technique. Without the template, the nucleic acids were observed at random locations on the nanoparticles. With the template, the two oligonucleotides were always seen at distinct geometric positions as arrangements controlled by the specific DNA nanostructure.
Suzuki says that top-down methods such as immobilization by a tip of scanning probe microscope are very precise, but prohibitively slow. In contrast, his team’s DNA template is extremely fast and automated, and represents a new type of ‘nanomachine.’
“Each nanomachine catches a certain number of oligonucleotides, immobilizes them onto a nanoparticle, and then releases them,” explains Suzuki. “Naturally, this task is best suited to a DNA template having complementary sequences to the oligonucleotides, since duplex formation is then completely reversible.”
According to Suzuki, creating nanoparticles with atom-like binding capabilities would have advantages beyond developing new types of nanostructures. “I knew that such a result would be welcomed by many other researchers and would accelerate the whole field,” he says.
*Suzuki, K., Hosokawa, K. & Maeda, M. Controlling the number and positions of oligonucleotides on gold nanoparticle surfaces. Journal of the American Chemical Society 131, 7518–7519 (2009).