Drugs based on engineered proteins represent a new frontier for pharmaceutical makers. Even after they discover a protein that may form the basis of the next wonder drug, however, they have to confront a long-standing problem: how to produce large quantities of the protein in a highly pure state. Now, a multi-institutional research team including a biochemist at the National Institute of Standards and Technology (NIST) may have found* a new solution in an enzymatic "food processor" they can activate at will.
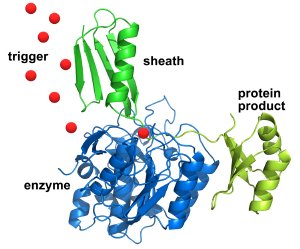 The sheath (in green at top), with the desired protein linked to it (in yellow at right), first sticks to the engineered enzyme (in blue atbottom left) – allowing impurities and other proteins to be washed away. Then an infusion of triggering molecules (red dots) causes the enzyme to cut away the desired protein (at the point where the sheath and protein connect), which can be collected in a pure state. The sheath can then be washed away and the process repeated. Credit: NIST
The sheath (in green at top), with the desired protein linked to it (in yellow at right), first sticks to the engineered enzyme (in blue atbottom left) – allowing impurities and other proteins to be washed away. Then an infusion of triggering molecules (red dots) causes the enzyme to cut away the desired protein (at the point where the sheath and protein connect), which can be collected in a pure state. The sheath can then be washed away and the process repeated. Credit: NIST
The team has found an efficient method of harvesting purified protein molecules by altering an enzyme that soil bacteria use to break down their food. In its natural form, this enzyme would be of little use to drug developers, but the team has modified it so that it can be activated at the desired moment. By creating essentially an "on-switch" for the enzyme's activity, the team has found a way to separate a single, desired protein from the mixture of thousands generated by a living cell, which remains biotechnology's natural protein factory of choice.
Bacteria use the enzyme, called subtilisin, as a sort of food processor: After producing it internally, they release the enzyme into the soil, where it uses a minuscule "blade" to chop up proteins into digestible pieces. Because it could damage the bacterium's interior, the blade has a protective sheath that only comes off once the enzyme has exited the cell.
"The enzyme and sheath are strongly attracted to each other. The enzyme's first act is to cut the sheath away," says NIST's Travis Gallagher. "The method takes advantage of their attraction in order to isolate the protein we want."
The team first creates many "sheathless" copies of the enzyme, which are modified to function only in the presence of a triggering molecule such as fluoride. The modified enzymes are bound to the surface of a strainer. Then the team uses engineered cells to generate mass quantities of a potentially therapeutic protein, each copy of which has a subtilisin sheath attached to it. After harvesting these proteins along with the thousands of others that grow in the cellular interior, they filter the mixture through the strainer, where the protein-sheath pairs are caught and stuck fast to the subtilisin while the rest of the mixture drains away.
At this point, the team flicks their switch. They add a bit of fluoride and the enzyme snips the bond between sheath and protein, releasing the desired protein free of almost all impurities. "The technique can conceivably be used to obtain any protein you like, and the process is repeatable, as the sheaths can be removed for another round of purification," Gallagher says. "For most proteins, the method can achieve greater than 95 percent purity at a single step."
The research team also includes members from Potomac Affinity Proteins, LLC (PAP) and the University of Maryland Biotechnology Institute (UMBI). UMBI holds the patent, which has been licensed to PAP. The research was supported by grants from the National Institutes of Health and the Bill and Melinda Gates Foundation.
* T. Gallagher, B. Ruan, M. London, M. Bryan and P.N. Bryan. Structure of a switchable subtilisin complexed with substrate and with the activator azide. Biochemistry, DATE, pages, DOI.