A research team at the University of California, Riverside has created a computer programming language that will automate “laboratory-on-a-chip” technologies used in DNA sequencing, drug discovery, virus detection and other biomedical applications.
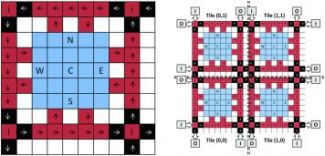 A tile is the fundamental building block of the virtual architecture. The virtual architecture is imposed onto a Digital Microfluidic BioChip by tiling the fundamental building blocks to create a 2D array of tiles.
A tile is the fundamental building block of the virtual architecture. The virtual architecture is imposed onto a Digital Microfluidic BioChip by tiling the fundamental building blocks to create a 2D array of tiles.
A laboratory-on-a-chip is a device that integrates laboratory functions on a chip that is only millimeters or centimeters in size. The technology allows for the automation and miniaturization of biochemical reactions. It has the potential and to improve and reduce the cost of healthcare.
“If you think of the beginning of computers they were basically tools to automate mathematics,” said Philip Brisk, an assistant professor in the Department of Computer Science and Engineering at UC Riverside’s Bourns College of Engineering. “What are we are creating is devices that could automate chemistry in much the same way.”
The most recent laboratory-on-a-chip devices are equipped with integrated electronic sensors, similar in principle to those used in today’s smart phones and tablet PCs. These sensors enable scientists and health care professionals working with the devices to analyze the sensor data to make informed decisions about future analyses to perform.
Brisk and his research team are funneling the sensor data into a computer, facilitating automated decision making, rather than employing a human-in-the-loop.
“We are really trying to eliminate as much human interaction as possible,” Brisk said. “Now, you have a chip, you use it and then you analyze it. Through automation and programmability, you eliminate human error, cuts costs and speed up the entire process.”
Brisk’s findings were recently published in a paper, “Interpreting Assays with Control Flow on Digital Microfluidic Biochips,” in ACM Journal on Emerging Technologies in Computing Systems.
There were two co-authors: Daniel Grissom, one of Brisk’s Ph.D. students; and Christopher Curtis, who worked with Brisk for three years as an undergraduate and plans to return as a Ph.D. student in the fall.
The team started with an existing biological programming language, BioCoder, developed by Microsoft’s research office in India. It was originally created to improve the reproducibility and automation of biology experiments by using a programming language to express the series of steps taken.
The UC Riverside team modified BioCoder to process sensor feedback in real-time. Using a software simulator to mimic the behavior of a laboratory-on-a-chip, they proved it works. Now, in conjunction with William Grover, an assistant professor of bioengineering at UC Riverside, they plan to build a prototype chip that can be used for real world applications.
This research is supported by the National Science Foundation under grant CNS-1035603, an NSF Graduate Research Fellowship awarded to Grissom, and a UC Riverside Dissertation Year Fellowship, also awarded to Grissom, who completes his Ph.D. in June, 2014.
Brisk was also recently awarded a five-year, $493,645 National Science Foundation CAREER grant for related research to apply semiconductor design automation and layout principles to laboratory-on-a-chip technology.