Feb 3 2016
Scientists at the Universities of York and Torino have used mathematics as a tool to provide precise details of the structure of protein nanoparticles, potentially making them more useful in vaccine design.
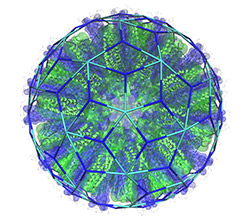 The picture shows a model of one of the SAPN particles that has been constructed by Newton Wahome based on a tiling by Giuliana Indelicato, illustrating how tiling theory predicts the surface architecture of the nanoparticle. Wahome and Indelicato are the joint first authors of this paper.
The picture shows a model of one of the SAPN particles that has been constructed by Newton Wahome based on a tiling by Giuliana Indelicato, illustrating how tiling theory predicts the surface architecture of the nanoparticle. Wahome and Indelicato are the joint first authors of this paper.
Working with a world-leading group at the University of Connecticut in the USA, who pioneered the development of self-assembling protein nanoparticles (SAPNs) for vaccine design, they have used advanced mathematical calculations to create a complete picture of the surface morphology of these particles. The research is published in the Biophysical Journal.
The nanoparticles self-assemble symmetrically using protein building blocks to create cage or shell-like architectures, which serve a range of functions such as storage, catalysis and structural scaffolding, or as enclosures for viral genomes. But electron microscopy and neutron scattering data has limited effectiveness for researchers attempting to classify the morphology of the nanoparticles.
Using mathematics to predict the geometries of nanoparticles can help scientists to select those whose structures are the most advantageous for the design of new vaccines. The constant need for vaccine development as new strains of disease evolve has generated a world market worth $56 billion a year.
The new study focused on a class of artificial SAPNs designed by Professor Peter Burkhard, a structural biophysicist at the University of Connecticut. When chemically attached to antigens from pathogens, nanoparticles can create simple, potent and cost-effective vaccines. Clinical tests on a malaria vaccine designed in this way are due to start soon.
Researchers at York and Torino, led by biophysicist Professor Reidun Twarock, of the University of York’s York Centre for Complex Systems Analysis and the Departments of Mathematics and Biology, used a mathematical tool called tiling theory to predict the symmetric classification of different particle morphologies of SAPNs. They adapted the tiling approach Professor Twarock previously pioneered in the context of virology to model protein nanoparticles with a mixture of local five- and three-fold symmetry axes.
Professor Twarock said: “We have developed a mathematical approach that allows you to identify the surface structures of these nanoparticles that you cannot get from experimentation alone. Mathematics plays an important role here because it acts like a microscope and helps to give researchers insights they couldn’t get experimentally.”
Professor Burkhard added: “The protein nanoparticles show great promise as future vaccine carriers and our malaria vaccine will be tested in a clinical setting within the next year. Understanding the geometric principles of the self-assembly to nanoparticles is essential for the successful design and development as vaccines.”