Mar 25 2016
c-myc mRNA has been established as a potential biomarker for detecting cancer, thanks to its critical function as a tumor suppressor or oncogene. Abnormal mRNA expression, in particular, is often seen during the development of early stage colon cancer. As a result, sensitive and specific detection of c-myc mRNA offers a potential way to make an early diagnosis of cancer, and has potential for the development of precision medicine.
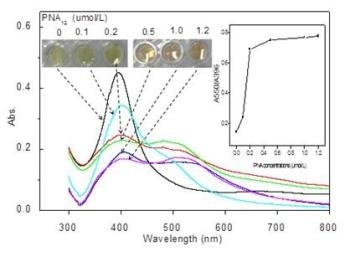 Depicts visual colors of AgNP solutions (0.45 nmol/L) incubated with different PNA12 concentrations (0-1.2 ìmol/L) and absorption spectra of corresponding solutions. (CREDIT: ©Science China Press)
Depicts visual colors of AgNP solutions (0.45 nmol/L) incubated with different PNA12 concentrations (0-1.2 ìmol/L) and absorption spectra of corresponding solutions. (CREDIT: ©Science China Press)
An international team of researchers has developed a label-free colorimetric method for sensitive detection of c-myc mRNA biomarkers. The method is based on peptide nucleic acid/silver nanoparticles (PNA/AgNPs).
The study article, titled "A label-free colorimetric assay for detection of c-myc mRNA based on peptide nucleic acid and silver nanoparticles" has been reported in the Science Bulletin journal. It was written by Dr. Xia Li from Liaocheng University (Shandong, China), Shandong Taishan Scholar Professor Jifeng Liu (current, Tianjin Science and Technology University) and Professor Chenzhong Li at the Florida International University (Miami, USA).
Just like DNA, PNA exhibits the same hydrogen-bonding nucleobases, except that it is bound to an uncharged N-(2-aminoethyl) glycine backbone that imparts physicochemical robustness and excellent target-binding properties over native DNA probes.
PNA can cause aggregation of AgNPs, reducing the optical absorbance and resulting in a change of color from yellow to red-brown. PND-mRNA complexes can be used to reversibly control AgNPs aggregation. To form these complexes, the complementary PNS probe-containing solution is added with c-myc mRNA. It is possible to view this reversible dissociation process by tracking the change in color from red-brown to yellow.
The China-U.S. collaborative team also studied the basic mechanism of molecular interactions between PNA and AgNPs. Through molecular modeling, the researchers examined the most likely arrangement of the PNA structure on AgNPs, and observed that different hydrogen bonds form between AgNPs and PNA, leading to natural aggregation of AgNPs with one another. They also believe that reduced electrostatic repulsion could play a role in PNA-induced AgNPs aggregation.
The PNA, in the presence of c-myc mRNA target, prefers to change its configuration to integrate with the target, complementary mRNA. This causes AgNPs to disaggregate into dispersed Ag nanoparticles.
This PNA and AgNPs integrated technology is quite simple, specific and sensitive. We are also considering transferring the liquid-based sensing platform to solid bio-sensing devices such as through a paper or silicon-based platform. In addition, we aim to integrate the bar code technology and smart phone to develop a portable oncogene screening biosensor for point-of-care applications.
Chenzhong Li, Professor of Biomedical Engineering, Florida International University
The National Natural Science Foundation of China (21305058, 21205056, 21075058 and 21503104) and Tai-Shan Scholar Research Fund of Shandong Province partily funded the study.