Oct 30 2007
More than 40 years after beta blockers were first used clinically, scientists can finally get a detailed, three-dimensional look at the drugs’ molecular target - the beta2-adrenergic receptor. This receptor hails from a family of proteins called G protein-coupled receptors (GPCRs) that control critical bodily functions, several of our senses, and the action of about half of today’s pharmaceuticals. Because this is the first known structure of a human GPCR, the work promises not only to speed the discovery of new and improved drugs, but also to broaden our understanding of human health and disease.
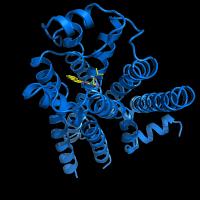 Crystal structure of the beta2-adrenergic receptor protein, the first known structure of a human G protein-coupled receptor.
Crystal structure of the beta2-adrenergic receptor protein, the first known structure of a human G protein-coupled receptor.
Published online in the October 25 issue of Science Express, the research was supported by two major initiatives of the National Institutes of Health—the NIH Roadmap for Medical Research and the Protein Structure Initiative (PSI), which is led by the National Institute of General Medical Sciences. Additional funding came from a Javits Neuroscience Award from the National Institute of Neurological Disorders and Stroke.
To solve the new structure, the researchers overcame daunting scientific obstacles. Many of these difficulties arise because GPCRs are membrane proteins—some of the trickiest molecules to capture in three-dimensional detail because they resist forming crystals, which are needed for structure determination.
“Because of their role in so many medically important processes and the great challenges they present for detailed study, membrane proteins have been one focus of the NIH Roadmap,” said NIH Director Elias A. Zerhouni, M.D. “The determination of this structure is an exciting example of the rewards from the Roadmap investment.”
Scientists across the country have tried for years to obtain detailed images of GPCR proteins. They’ve succeeded only once before, in 2000, when a research group determined the structure of a visual pigment in cow eyes. Part of the job was easier then, because the pigment protein is abundant. One of the major challenges in the new study was getting enough of the beta2-adrenergic receptor protein to work with.
After trying in vain to get the protein in its natural form to crystallize, Brian Kobilka and co-workers at Stanford University turned to protein engineering. To overcome problems with the protein’s floppiness, they replaced a flexible loop with a more rigid protein structure. Raymond Stevens and co-workers of The Scripps Research Institute managed to coax the intransigent proteins into tiny crystals by devising techniques and environmental conditions that mimicked the protein’s native, membrane-like environment. These genetic engineering and crystallization techniques may be broadly applicable to similar proteins, paving the way for more structures from the hundreds of GPCRs encoded in the human genome.
“This is an absolutely remarkable advance,” said Jeremy M. Berg, Ph.D., director of NIGMS, which, in addition to spearheading the PSI, plays a leading role in the membrane protein Roadmap initiative. “Many laboratories around the world are trying to reveal the secrets of these proteins, and this important structure takes the field to a higher level.”