Jun 25 2009
In a landmark technical achievement, investigators in the Vanderbilt Center for Structural Biology have used nuclear magnetic resonance (NMR) methods to determine the structure of the largest membrane-spanning protein to date.
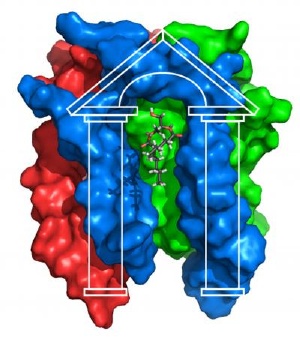 Surface-filled representation of diacylglycerol kinase. The "porch-like" structure of the enzyme is highlighted, and the substrate diacylglycerol is depicted bound to the active site. Investigators at the Vanderbilt Center for Structural Biology used NMR methods to determine the structure of diacylglycerol kinase, the largest membrane-spanning protein studied by NMR to date. Credit: Charles Sanders, Ph.D., Vanderbilt University Center for Structural Biology
Surface-filled representation of diacylglycerol kinase. The "porch-like" structure of the enzyme is highlighted, and the substrate diacylglycerol is depicted bound to the active site. Investigators at the Vanderbilt Center for Structural Biology used NMR methods to determine the structure of diacylglycerol kinase, the largest membrane-spanning protein studied by NMR to date. Credit: Charles Sanders, Ph.D., Vanderbilt University Center for Structural Biology
Although NMR methods are routinely used to "take molecular pictures" of small proteins, large proteins – and particularly those that reside within the cell membrane – have been reluctant to smile for the camera.
In the June 26 issue of Science, Charles Sanders, Ph.D., professor of Biochemistry, and colleagues report the NMR structure of the large bacterial protein diacylglycerol kinase (DAGK), a complex of three subunits that each cross the membrane three times (for a total of nine membrane spans).
The group's ability to determine the NMR structure of DAGK suggests that similar methods can now be used to study the structures of other membrane proteins.
"We're taking the methods that we used for diacylglycerol kinase and applying them to high value targets such as G protein-coupled receptors," Sanders said.
G protein-coupled receptors – the largest family of cell signaling proteins – are targets for about half of all pharmaceuticals. Sanders is collaborating with other Vanderbilt investigators to tackle G protein-coupled receptor structure using both NMR and a complementary structural approach, X-ray crystallography.
DAGK may be a therapeutic target for certain types of bacterial infections. It is a virulence factor in the bacteria Streptococcus mutans, which causes tooth decay.
Sanders selected DAGK as a model for studying membrane enzymes when he started his own research lab 17 years ago. DAGK is the smallest known kinase (a protein that adds chemical groups called phosphates onto other molecules), and it is not similar to any other known proteins.
The DAGK structure, Sanders said, "confirmed that this is a really strange kinase." The enzyme has a porch-like structure, with a wide opening for its substrate diacylglycerol and the active site at the top of the porch.
"The active site looks nothing like any other kinase active site – it's a unique architecture," Sanders said.
The researchers also performed exhaustive mutagenesis studies in which they characterized mutations at each amino acid in DAGK and used the data to map the active site of the enzyme onto the structure. They identified two sets of mutations that resulted in non-functional DAGK. One set altered the active site so that it no longer did its job, and the second set caused the protein to fold incorrectly (misfolding).
Sanders said the team was surprised to find that nearly all of the mutations that caused misfolding were in the active site. The expectation, he explained, is that mutations in the active site would cause a loss of function but would not usually affect protein folding, whereas key residues for folding would be located elsewhere in the protein to underpin the scaffold for the active site.
"Our study shows that you can't make that assumption," he said.
Sanders cautions that investigators cannot simply predict the impact of a mutation based on it being located in the active site. The finding has implications for personalized medicine, which aims to use the predicted impact of disease-causing mutations to make therapy decisions.
"The therapeutic strategy for addressing catastrophic misfolding versus simple loss of function may be very different," Sanders said.
Sanders and his team, who got interested in protein folding because of their work with DAGK, are now pursuing structural studies of misfolded membrane proteins that cause diseases including peripheral neuropathy (Charcot-Marie-Tooth Disease), diabetes insipidus and Alzheimer's disease.
"For proteins that misfold because of mutations, we're using NMR tools to understand exactly what the mutations do to the proteins in terms of structure and stability," Sanders said. "We believe that understanding will lead to predictions about how to intervene and avoid misfolding."
Co-authors include Wade Van Horn, Ph.D., Hak-Jun Kim, Ph.D., Charles Ellis, Ph.D., Arina Hadziselimovic, Endah Sulistijo, Ph.D., Murthy Karra, Ph.D., and Changlin Tian, Ph.D., at Vanderbilt and Frank Sönnichsen, Ph.D., at Christian Albrechts University in Kiel, Germany.
Sanders said the resources and collegial environment of the Department of Biochemistry and the Vanderbilt Center for Structural Biology, which he joined seven years ago, made it possible for his team to complete the DAGK structure. The National Institutes of Health supported the research.