Jun 2 2010
Since the 1990s, a green fluorescent protein known simply as GFP has revolutionized cell biology. Originally found in a Pacific Northwest jellyfish, GFP allows scientists to visualize proteins inside of cells and track them as they go about their business. Two years ago, biologists who discovered and developed the protein as a laboratory tool won a Nobel Prize for their work.
However, using GFP as a fluorescent probe has one major drawback — the protein is so bulky that it can interfere with the proteins it's labeling, preventing them from doing their normal tasks or reaching their intended destinations.
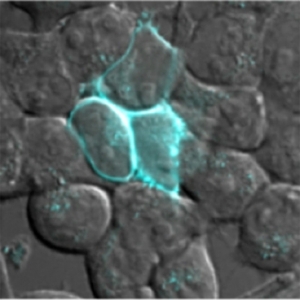 MIT researchers have designed a fluorescent probe that can be targeted to different locations within a cell.
MIT researchers have designed a fluorescent probe that can be targeted to different locations within a cell.
"For a long time, people have been trying to find better ways to label proteins," says Katharine White, an MIT graduate student in the lab of Alice Ting, associate professor of chemistry.
Ting, White and their colleagues have now come up with a new way to overcome the disadvantages of GFP, by tagging proteins with a much smaller probe. Their probe allows proteins to carry out their normal functions, offering scientists the chance to glimpse never-before-seen activity.
The researchers describe the new technique, dubbed PRIME (PRobe Incorporation Mediated by Enzymes), in the Proceedings of the National Academy of Sciences this week.
First isolated from a jellyfish in 1962, GFP allows scientists to track otherwise invisible proteins as they move about the cell, orchestrating processes such as cell division and metabolism. To achieve this, scientists tack the gene for GFP onto the gene for the protein they want to study. After the engineered gene is introduced into cells, it will produce proteins that glow fluorescent green.
However, GFP's large size (238 amino acids) can interfere with some proteins, such as actin, a molecule that helps give cells their structure and is involved in cell division, motility and communication with other cells.
"People use fluorescent proteins to study actin all the time, but fusion to the fluorescent proteins has detrimental effects on actin's function and trafficking," says Tao Uttamapinant, co-lead author of the PNAS paper with White and former MIT postdoctoral associate Hemanta Baruah.
To overcome the drawbacks of GFP, Ting and her students used a blue fluorescent probe that is much smaller than GFP. Unlike GFP, the new probe is not joined to the target protein as it's produced inside the cell. Instead, the probe is attached later on by a new enzyme that the researchers also designed.
For this to work, the researchers must add the gene for the new enzyme, known as a fluorophore ligase, to each cell at the same time that they add the gene for the protein of interest. They also add a short tag (13 amino acids) to the target protein, and this tag allows the enzyme to recognize the protein. When the blue fluorescent probe (7-hydroxycoumarin) is added to the cell, the enzyme attaches it to the short tag on the target protein.
With this method, proteins such as actin can move freely throughout the cell and cross into the nucleus even when tagged with the fluorescent probe.
The researchers also demonstrated that they can label proteins in specific parts of the cell, such as the nucleus, cell membrane or cytosol (the interior of the cell), by tagging the enzyme with genetic sequences that direct it to specific locations. That way, the enzyme attaches the fluorescent probe only to proteins in those locations.
The MIT team is now working on engineering enzymes that will work with other types of probes. Ting has also filed for a patent on the fluorescent probe technique and plans to commercialize the technology so other labs can use it.
Source: http://web.mit.edu/