Scientists at The Scripps Research Institute (TSRI) have made a major advance in understanding how flu viruses replicate within infected cells. The researchers used cutting-edge molecular biology and electron-microscopy techniques to “see” one of influenza’s essential protein complexes in unprecedented detail. The images generated in the study show flu virus proteins in the act of self-replication, highlighting the virus’s vulnerabilities that are sure to be of interest to drug developers.
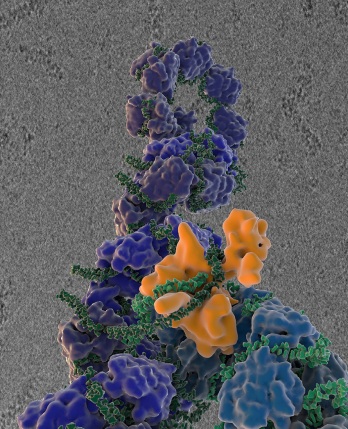 The new Scripps Research Institute study shows flu virus proteins in the act of self-replication. Shown here is the influenza virus, which encapsidates its RNA genome (green) with a viral nucleoprotein (blue); the influenza virus polymerase (orange) reads and copies the RNA genome. In the background is an image of influenza virus ribonucleoprotein complexes observed using cryo-electron microscopy. (Image courtesy of the Wilson, Carragher and Potter labs.)
The new Scripps Research Institute study shows flu virus proteins in the act of self-replication. Shown here is the influenza virus, which encapsidates its RNA genome (green) with a viral nucleoprotein (blue); the influenza virus polymerase (orange) reads and copies the RNA genome. In the background is an image of influenza virus ribonucleoprotein complexes observed using cryo-electron microscopy. (Image courtesy of the Wilson, Carragher and Potter labs.)
The report, which appears online in Science Express on November 22, 2012, focuses on influenza’s ribonucleoprotein (RNP). RNPs contain the virus’s genetic material plus the special enzyme that the virus needs to make copies of itself.
“Structural studies in this area had stalled because of the technical obstacles involved, and so this is a welcome advance,” said Ian A. Wilson, the Hansen Professor of Structural Biology at TSRI and senior author of the report with TSRI Professors of Cell Biology Bridget Carragher and Clint Potter. “The data from this study give us a much clearer picture of the flu virus replication machinery.”
Unveiling the Mystery of RNPs
At the core of any influenza virus lie eight RNPs, tiny molecular machines that are vital to the virus’s ability to survive and spread in its hosts. Each RNP contains a segment—usually a single protein-coding gene—of the RNA-based viral genome. This viral RNA segment is coated with protective viral nucleoproteins and has a structure that resembles a twisted loop of chain. The free ends of this twisted loop are held by a flu-virus polymerase enzyme, which handles the two central tasks of viral reproduction: making new viral genomic RNA, and making the RNA gene-transcripts that will become new viral proteins.
Aside from its importance in ordinary infections, the flu polymerase contains some of the key “species barriers” that keep, for example, avian flu viruses from infecting mammals. Mutations at key points on the enzyme have enabled the virus to infect new species in the past. Thus researchers are eager to know the precise details of how the flu polymerase and the rest of the RNP interact.
Getting those details has been a real challenge. One reason is that flu RNPs are complex assemblies that are hard to produce efficiently in the lab. Flu polymerase genes are particularly resistant to being expressed in test cells, and their protein products exist in three separate pieces, or subunits, that have to somehow self-assemble. Until now, the only flu RNPs that have been reproduced in the laboratory are shortened versions whose structures aren’t quite the same as those of native flu RNPs. Researchers also are limited in how much virus they can use for such studies.
The team nevertheless managed to develop a test-cell expression system that produced all of the protein and RNA components needed to make full-length flu RNPs. “We were able to get the cells to assemble these components properly so that we had working, self-replicating RNPs,” said Robert N. Kirchdoerfer, a first author of the study. Kirchdoerfer was a PhD candidate in the Wilson laboratory during the study, and is now a postdoctoral research associate in the laboratory of TSRI Professor Erica Ollmann Saphire.
Kirchdoerfer eventually purified enough of these flu RNPs for electron microscope analysis at TSRI’s Automated Molecular Imaging Group, which is run jointly by Carragher and Potter.
Never Seen Before
The imaging group’s innovations enable researchers to analyze molecular samples more easily, in less time, and often with less starting material. “We were able, for example, to automatically collect data for several days in a row, which is unusual in electron microscopy work,” said Arne Moeller, a postdoctoral research associate at the imaging group who was the other first author of the study.
Electron microscopes make high-resolution images of their tiny targets by hitting them with electrons rather than photons of light. The images revealed numerous well-defined RNP complexes. To Moeller and his colleagues’ surprise, many of these appeared to have new, partial RNPs growing out of them. “They were branching—this was very exciting,” he said.
“Essentially these were snapshots of flu RNPs being replicated, which had never been seen before,” said Kirchdoerfer. These and other data, built up from images of tens of thousands of individual RNPs, allowed the team to put together the most complete model yet for flu-RNP structure and functions. The model includes details of how the viral polymerase binds to its RNA, how it accomplishes the tricky task of viral gene transcription, and how a separate copy of the viral polymerase assists in carrying out RNP replication. “We’re now able to take a lot of what we knew before about flu virus RNP and map it onto specific parts of the RNP structure,” said Kirchdoerfer.
The new flu RNP model highlights some viral weak points. One is a shape-change that a polymerase subunit—which grabs viral RNA and feeds it to the polymerase’s active site on a second subunit—has to undergo during viral gene transcription. Another is key interaction between the polymerase and viral nucleoproteins. Flu RNPs are long and flexible, curving and bending in electron microscope images; and thus the structural model remains only modestly fine-grained. “You wouldn’t be able to design drugs based on this model alone,” said Kirchdoerfer, “but we now have a much better idea of how flu RNPs work, and that does suggest some possibilities for better flu drugs.”
The study, “Organization of the Influenza Virus Replication Machinery,” was funded in part by grants from the National Institutes of Health (AI058113, GM095573) and the Joint Center for Innovation in Membrane Protein Production for Structure Determination (P50GM073197). TSRI’s Automated Molecular Imaging Group includes the National Resource for Automated Molecular Microscopy, which is supported by the National Institutes of Health’s National Center for Research Resources (2P41RR017573-11) and the National Institute of General Medical Sciences Biomedical Technology Resource Centers (9 P41 GM103310-11).