Sep 1 2014
Cancer patients may soon receive medication more efficiently and with fewer harmful side effects thanks to a new development in nanofiber assembly.
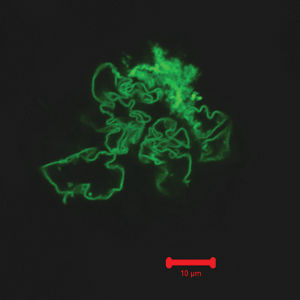 Method for building larger nanofibers discovered, Researchers discovered a new method for creating large synthetic protein fibers. Photo courtesy of CMU
Method for building larger nanofibers discovered, Researchers discovered a new method for creating large synthetic protein fibers. Photo courtesy of CMU
Researchers at the University of Pittsburgh, Carnegie Mellon University and Oregon State University have discovered a method for creating large synthetic fibers made of proteins. These structures could provide the key for future research in medicine and pharmacology.
The protein nanofibers could provide a more effective means for delivering anticancer drugs to tumors within the body. By attaching to the surface of tumor cells, nanofibers could deliver the drug to those cells, leaving them unaffected.
The study, “Cooperative, Reversible Self-Assembly of Covalently Pre-Linked Proteins into Giant Fibrous Structures,” details how and why green fluorescent proteins form into massive fibers.
The proteins, which emit a green fluorescence under ultraviolet light, build biological structures outside of cells. Saadyah Averick, a chemistry graduate student at CMU and head of the research project, wanted to connect these proteins into massive nanofibers, millimeters in length. This would allow larger biological structures to be constructed.
To make the proteins able to self-assemble, he sought help from Dr. Ryan Mehl, an associate professor of biochemistry at Oregon State University. Mehl modified the amino acids, the building blocks of the protein, and gave each protein two heads to attach to other proteins. When Averick modified a polymer that could link to the protein in the same way and attached it, the proteins self-assembled into long chains.
“Now, [Averick] could make a polymer with two heads that will react with the protein,” Mehl said in an email. “Since the polymer and the protein both have two heads each that will only react with each other (like Velcro), they zip together into wires or fibers.”
The research and report were joint efforts between Pitt and CMU and included CMU’s Dr. Krzysztof Matyjaszewski and Dr. Tomasz Kowalewski, professors in CMU’s chemistry department, and Pitt’s Dr. Anna C. Balazs — all leading researchers in chemistry and chemical engineering. For the team, the discovery was serendipitous.
“From short chains [of the molecules], we can form large uniform fibers, something unexpected,” Averick said.
According to the study, the self-assembly of the proteins can be reversed. The team observed that the fibers broke apart into their original, small pieces when exposed to sound waves. Several days later, the fibers reassembled. This property makes the fibers ideal for drug delivery systems, as it allows them to be injected into the bloodstream. Injection into the bloodstream is not new to this method but it is unique as it allows larger fibers to be injected, in comparison to other drug delivery systems.
Averick said he was originally working toward creating large proteins made from giant fibrous structures and discovered the nanofibers by accident.
“When I did the reaction, a soup of green fibers was obtained,” Averick said. “It was very frustrating. I tried many reactions to avoid the fiber formation but always found that [type of] fibers in my reaction mixture.”
Shortly after starting work on his doctorate at CMU in 2009, Averick began his work with proteins, modified by Mehl, and originally attempted to create short chains. After making the short chains, Averick said he hoped to join them all together to make large chains but was unsuccessful. He was left with a pool of green fibers.
“The fiber formation was very strange, since we only expected short [protein] chains to form,” Averick said.
For help, Averick said he turned to Kowalewski. Over the next 3 1/2 years, the two tried to analyze the green fibers using atomic force microscopy — an imaging technique — but never yielded good images. One day, they accidentally changed the components of a part of their sample and were able to get the first clear images of the fibers. They realized that they had the large protein chains they’d sought.
With this success, Kowalewski asked Balazs to join the research team.
Balazs created a computer model of the self-assembling proteins to study the fibers’ structure and their mechanism of self-assembly.
Although research is still in the developmental phase, Balazs said understanding how the proteins self-assemble could lead to fiber structures being built by the molecules they are made of. In tissue engineering, such as in reconstructive surgeries, this could mean much less invasive procedures.
“We will continue to work with the CMU team to design soft materials with controllable structures and new functionalities,” Balazs said in an email.
Angewandte Chemie International Edition, an English language chemistry journal published by the German Chemical Society, published the report in July 2014. This discovery is a branch off of Saadyah Averick’s earlier work on construction with proteins.
“This was purely curiosity-driven and serendipity-driven work,” Kowalewski said in a press release from the Mellon College of Science at CMU. “But where controlled polymerization and organic chemistry meet biology, interesting things can happen.”