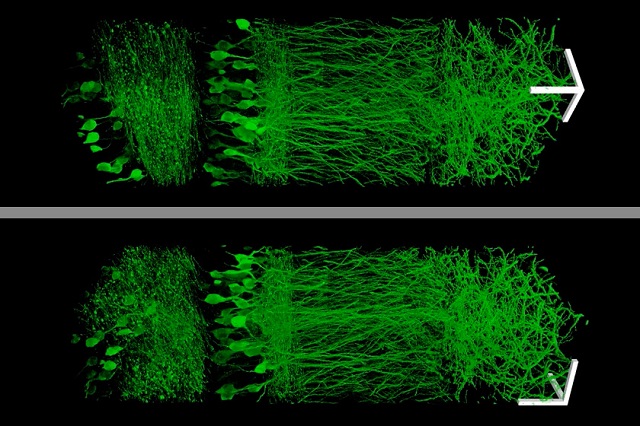
 Using a new technique that allows them to enlarge brain tissue, MIT scientists created these images of neurons in the hippocampus. (Courtesy: Fei Chen and Paul Tillberg)
Using a new technique that allows them to enlarge brain tissue, MIT scientists created these images of neurons in the hippocampus. (Courtesy: Fei Chen and Paul Tillberg)
Researchers from MIT have successfully discovered a technique to physically magnify tissue samples by implanting them in a polymer, which swells on addition with water to enable high-resolution images. Since the late 1500s when the first microscope was invented, scientists have been devising ways to study preserved tissues and cells in a better manner.
With the advent of the “super-resolution” microscopes, scientists now have a tool that provides a resolution which is better than 250nm while viewing inside cells. Instead of finding ways to improve the existing microscopes, the MIT team focused on ways to enlarge the tissue samples.
Expansion Microscopy Explained
The technique only required inexpensive microscopes and commonly available chemicals that can be found in most research labs. Thus this super-resolution imaging technique can be easily adapted by other researchers as well.
Instead of acquiring a new microscope to take images with nanoscale resolution, you can take the images on a regular microscope. You physically make the sample bigger, rather than trying to magnify the rays of light that are emitted by the sample.
Ed Boyden, an associate professor of biological engineering and brain and cognitive sciences at MIT
Generally, microscopes require the use of lenses to focus light transmitted from a sample into a magnified image. There is however a drawback with this method known as diffraction limit. The diffraction limit does not allow visualization of objects smaller than the wavelength of the light being used. For instance, if blue-green light is used with a 500nm wavelength, it will not be possible to view anything smaller than 250nm.
Unfortunately, in biology that’s right where things get interesting.
Boyden, a member of MIT’s Media Lab and McGovern Institute for Brain Research
Molecules transferring payloads in and out of cells, protein complexes, and other cellular actions are said to be ordered at the nanoscale.
Scientists have come up with some “really clever tricks” to overcome this limitation, Boyden says. However, these super-resolution processes ideally work well with small, thin samples, and require plenty of time to image large samples.
If you want to map the brain, or understand how cancer cells are organized in a metastasizing tumor, or how immune cells are configured in an autoimmune attack, you have to look at a large piece of tissue with nanoscale precision.
Boyden
Therefore to accomplish this, the MIT team chose to spend time working with the sample rather than the microscope. They embedded the samples in expandable polymer gel comprising polyacrylate, which is a highly absorbent material normally used in diapers.
The researchers labeled the cell parts which had to be analyzed with the help of an antibody. The antibody had the tendency to bind itself the selected targets, and it can be connected to a fluorescent dye and a chemical anchor, which can fasten the dye to the polyacrylate chain.
After labeling of the tissue, the researchers added the precursor to the polyacrylate gel and then heated it to create a gel. The proteins, holding the sample together, are digested, thereby permitting uniform expansion. The sample is then washed using salt-free water so as to stimulate the volume to expand a 100-times.
Despite the fact the proteins are now broken apart, the location of fluorescent labels remain the same in relation to the whole tissue structure as it is anchored to the polyacrylate gel.
What you’re left with is a three-dimensional, fluorescent cast of the original material. And the cast itself is swollen, unimpeded by the original biological structure.
Tillberg
On expanding the tissue sample, the MIT team imaged it using confocal microscopes, which is generally used for fluorescent imaging with a resolution limit of hundreds of nanometers. The researchers managed to achieve a resolution of 70nm with their enlarged samples.
The expansion microscopy process … should be compatible with many existing microscope designs and systems already in laboratories.
Chen
The MIT team then applied this newly discovered technique to image a portion of brain tissue 500 by 200 by 100µm with a standard confocal microscope.
Basically imaging large samples like brain tissue is challenging with other super resolution techniques since these require minutes for imaging a one micron-thick tissue slice and is restricted in its capability of large image sampling by other aberrations and optical sampling.
“The exciting part is that this approach can acquire data at the same high speed per pixel as conventional microscopy, contrary to most other methods that beat the diffraction limit for microscopy, which can be 1,000 times slower per pixel,” says George Church, a professor of genetics at Harvard Medical School who was not part of the research team.
“The other methods currently have better resolution, but are harder to use, or slower,” Tillberg says. “The benefits of our method are the ease of use and, more importantly, compatibility with large volumes, which is challenging with existing technologies.”
The researchers believe that their technique will prove to be highly useful to scientists focused on imaging brain cells and mapping ways to communicate with each other across large expanses.
“There are lots of biological questions where you have to understand a large structure,” Boyden says. “Especially for the brain, you have to be able to image a large volume of tissue, but also to see where all the nanoscale components are.”
Although the focus of the MIT team has been on brain tissue study, other probable applications for their innovative technique include visualizing how immune cells attack specific organs during autoimmune disease, or analyzing tumor metastasis and angiogenesis.
This MIT research was funded by the National Institutes of Health, Jeremy and Joyce Wertheimer, the New York Stem Cell Foundation, the Fannie and John Hertz Foundation, and the National Science Foundation.
References