May 4 2015
Scientists require high-resolution imaging of plant cells to study everything from fungal infections to reproduction in maize. These images are captured with scanning electron microscopy (SEM), where an electron microscope focuses beams of electrons to increase magnification of objects. SEM is a common technique for all fields of science.
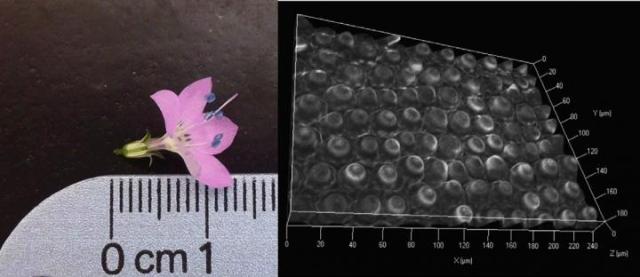 A mature flower of Saltugilia caruifolia (left), accompanied with a microscopic view of the petal lobes (right) showing the 3-D cell structure, which has implications in the flower's development and its ability to attract pollinators. Credit: Photo by Jacob B. Landis from: Landis, J. B., K. L. Ventura, D. E. Soltis, P. S. Soltis, and D. G. Oppenheimer. 2015. Optical sectioning and 3D reconstructions as an alternative to scanning electron microscopy for analysis of cell shape. Applications in Plant Sciences 3(4): 1400112. doi:10.3732/apps.1400112
A mature flower of Saltugilia caruifolia (left), accompanied with a microscopic view of the petal lobes (right) showing the 3-D cell structure, which has implications in the flower's development and its ability to attract pollinators. Credit: Photo by Jacob B. Landis from: Landis, J. B., K. L. Ventura, D. E. Soltis, P. S. Soltis, and D. G. Oppenheimer. 2015. Optical sectioning and 3D reconstructions as an alternative to scanning electron microscopy for analysis of cell shape. Applications in Plant Sciences 3(4): 1400112. doi:10.3732/apps.1400112
However, preparing objects for SEM and other common imaging methods can compromise delicate biological samples. Freeze-drying of material, electron beams, and vacuum pressure in the microscope can all result in cell damage, which is problematic for visualizing cell shape, size, and development. In addition, preparing material to be viewed under the microscope using SEM requires specialized equipment, causing a low throughput and placing a large financial burden on scientific labs.
Researchers at the University of Florida have developed new methods for a less damaging, more cost-effective, and higher-throughput approach for imaging cellular structure. "For visualizing cells of a variety of plant material," explains researcher Jacob Landis, "there are alternatives to the commonly used SEM. Some of these alternatives may be more easily accessible to researchers and do not require specialized equipment for processing prior to imaging."
Landis and colleagues created protocols for an optical sectioning-3D reconstruction method that uses a compound fluorescence light microscope. A compound microscope has multiple lenses to reflect light and project a high-magnification image. The microscope and a mounted camera "sections" the plant cells, capturing pictures of several layers and planes of the sample material. The images are reconstructed into a final 3D image with equal to higher resolution as images produced using SEM.
Landis developed the new protocol (detailed in a recent issue of Applications in Plant Sciences [http://www.bioone.org/doi/pdf/10.3732/apps.1400112]) to see epidermal cells on the surfaces of flower petals. This group of cells reveals extensive information to geneticists, agronomists, and ecologists. Flower epidermal cells cause all types of cellular-level changes that can scale up to influence flower color and development, as well as ecological interactions such as a plant's success in attracting pollinators and avoiding insect enemies.
Unlike in SEM, material to be imaged using the new method gets preserved, allowing for clear imaging months after the collection of any samples. This is valuable when studies require bulk processing of samples. The material is then dehydrated in ethanol alcohol, dipped in a liquid to remove the waxy film that protects the outer layer of petal cells, and mounted in a fluorescent dye to help visualize details under the microscope.
The method was catalyzed by a larger study on the cellular and genetic basis of flower size in the woodland plant group Saltugilia (see Image). Saltugilia plants have a large range of flower sizes and host all types of pollinators--including hummingbirds, bees, and bee flies--while some host no pollinators at all. SEM could not be used to understand these diverse flowers because it is limited to samples 3-5 mm in size. The new optical sectioning-3D reconstruction approach allows for samples as large as 5 cm to be imaged before cutting petals and compromising petal cellular structure.
Optical-sectioning and 3D reconstruction have been informally tested on other plant material, including leaf cells and reproductive organs (stamens, anthers, and sepals). "We believe that any kind of tissue that can be mounted on a microscope slide will be possible to image," says Landis. "I have spent a lot of time using SEM and prepping samples, and optical sectioning with 3D reconstruction is by far the simplest and most cost-effective method I have ever used."