Jun 11 2018
What is the driving factor for engines to move and cells to live? All this boils down to an entity termed “free energy,” which is typically the energy that can be derived from any system to carry out useful work. Any lack of this available energy would render a machine idle or will lead to the eventual death of a living organism.
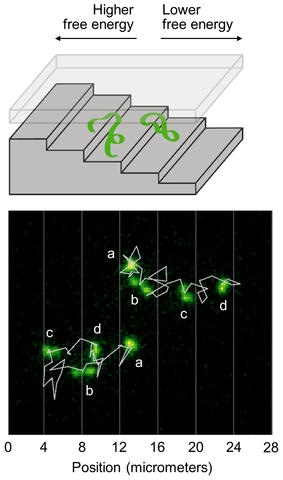 Illustration shows two DNA molecules in a nanofluidic staircase. The staircase confines the DNA molecules, creating a free energy that is higher at the top and lower at the bottom. The DNA molecules mostly descend the staircase to reduce their energy and relax, but sometimes ascend the staircase as microscopic fluctuations increase their energy. Bottom: Microscope images show two DNA molecules in the staircase. Jagged white lines show their trajectories. Letters mark different images of each molecule taken at 1-minute intervals. Vertical white lines show the positions of step edges. The molecule at the top right mostly descends the staircase. The molecule at the bottom left ascends two steps before descending. Relaxation Fluctuation Spectroscopy is a new method of analyzing such fluctuating trajectories to measure the free energy of microscopic systems. (Image credit: NIST)
Illustration shows two DNA molecules in a nanofluidic staircase. The staircase confines the DNA molecules, creating a free energy that is higher at the top and lower at the bottom. The DNA molecules mostly descend the staircase to reduce their energy and relax, but sometimes ascend the staircase as microscopic fluctuations increase their energy. Bottom: Microscope images show two DNA molecules in the staircase. Jagged white lines show their trajectories. Letters mark different images of each molecule taken at 1-minute intervals. Vertical white lines show the positions of step edges. The molecule at the top right mostly descends the staircase. The molecule at the bottom left ascends two steps before descending. Relaxation Fluctuation Spectroscopy is a new method of analyzing such fluctuating trajectories to measure the free energy of microscopic systems. (Image credit: NIST)
In a study at the National Institute of Standards and Technology (NIST) and the University of Maryland in College Park, an innovative way for measuring free energy has been developed and demonstrated by researchers. When compared to earlier techniques, the new technique can be applied to a wider range of nanoscopic and microscopic systems by employing microscopy for the tracking and analysis of the fluctuating motion or configuration of single molecules or other small objects.
Scientists have relied on free energy to understand complex systems since the development of steam engines. This concept will continue to be just as fundamental as we engineer and design proteins and other single-molecule systems. But the measurements are much harder for those small systems—so approaches like the new one we describe will be of fundamental importance.
David Ross, first author of a new paper on this study, NIST
Researchers can predict specific features related to the behavior of a living system or the operation of machine by measuring variations in free energy when a system moves or alters its internal structure, and this can be achieved without the need for the unmanageable task of keeping track of the entry and exit of all the atoms and molecules that constitute the system.
A day-to-day example of free energy is energy in an automobile internal combustion engine, where total energy is equal to the sum of the energy of its motion and the heat produced. The free energy can be obtained by subtracting the heat energy, which gets dissipated from the system.
Studies on the detailed behavior of microscopic systems — the realm of living cells, small molecules, and nanotechnologies — have shown that the measurement of variations in free energy is more challenging when compared to larger scales.
In one technique, researchers used a microscopic force sensor to pull on a DNA or protein molecule, which could function like a miniature spring when compressed or stretched, to measure the variations in position and force when a system relaxes and liberates energy. Yet, the microscopic system could be perturbed by the attachment of the force sensor and it would become impossible to use the sensor to measure variations in free energy that do not entail a direct change in position.
The innovative method, which enables the use of optical microscopy for tracking the motion or configuration of small systems, can be employed to determine free energies without the need for attaching a force sensor. The new study could turn out to be a robust way to gain insights into the intrinsic properties of a wide range of microscopic systems, such living systems like cells or viruses to better perceive the processes, such as chemical reactions, energy intake, and the movement of molecules that sustain the functioning of the living systems.
“We are surrounded by natural systems that take advantage of microscopic fluctuations in free energy, and now we have a way to better measure, understand, and, ultimately, manipulate these fluctuations ourselves,” stated Elizabeth Strychalski of NIST, a co-author of the study.
The analysis is quite suitable for investigating microscopic systems that start from a highly excited state with high energy, far from equilibrium with their surroundings, and then relax back toward equilibrium. The characteristics of microscopic systems can considerably vary when they relax owing to the random motion from continuous collision with surrounding molecules. In the new technique, named Relaxation Fluctuation Spectroscopy (ReFlucS) by the researchers, measurements of those fluctuations during relaxation are used to estimate the free energy.
Our approach shows that useful information can be gleaned from observing the random motions of a system as it settles down from a highly excited, far-from-equilibrium state.
Christopher Jarzynski, The University of Maryland, one of the co-authors of the study
As an example, the researchers investigated the motion of DNA molecules restricted to a nanometer-scale space with the shape of a staircase. To be restricted to the top steps, the shallowest, the DNA molecules have to be compressed more firmly when compared to molecules occupying the bottom steps. This leads to an increase in free energy for the molecules at the top. The researchers applied an electric field to drive the DNA molecules to the top of the staircase. Then, they turned off the electric field and analyzed the movement of the molecules using an optical microscope.
It was found that mostly, the DNA molecules descended the staircase when they relaxed toward equilibrium, leading to a decrease in their free energy. Yet, microscopic fluctuations induced the DNA molecules to occasionally move back up the staircase, resulting in an increase in their free energy. The fluctuating motion of the DNA molecules was investigated by the team, thereby allowing the free-energy profile — the amount of free energy at different locations, and the points where the energy is high and low — to be mapped out.
“ReFlucS provides access to information about free energy that was previously inaccessible,” stated Samuel Stavis from NIST, another co-author of the study.
Microscope video shows two DNA molecules in a nanofluidic staircase, with jagged white lines indicating their trajectories. Vertical white lines show the positions of step edges. The molecule at top right mostly descends the staircase. The molecule at bottom left ascends two steps before descending. Relaxation Fluctuation Spectroscopy is a new method of analyzing such fluctuating trajectories to measure the free energy of microscopic systems. (Credit: NIST)