Mar 13 2020
Removing antibiotic-resistant bacteria from wastewater is not enough to eliminate the risks posed by these organisms to society. These bacteria leave behind bits that had to be destroyed as well.
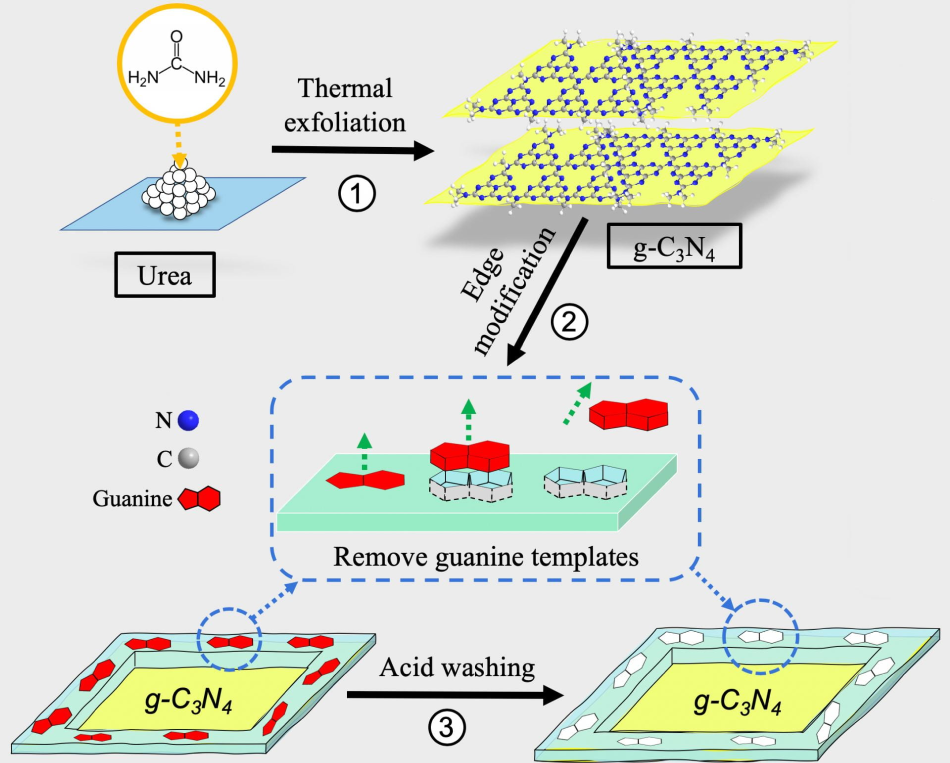 A schematic shows the three-step method to produce molecular-imprinted graphitic carbon nitride nanosheets. The process developed by Rice researchers could help catch and kill free-floating antibiotic-resistant genes found in secondary effluent produced by wastewater plants. Image Credit: Danning Zhang.
A schematic shows the three-step method to produce molecular-imprinted graphitic carbon nitride nanosheets. The process developed by Rice researchers could help catch and kill free-floating antibiotic-resistant genes found in secondary effluent produced by wastewater plants. Image Credit: Danning Zhang.
At Rice University’s Brown School of Engineering, scientists have developed a new method for “trapping and zapping” genes that are resistant to antibiotics. These genes are pieces of bacteria that, despite the death of their hosts, can still find their way and increase the resistance of other pathogens.
The research team, headed by Pedro Alvarez, an environmental engineer at Rice University, is utilizing molecular-imprinted graphitic carbon nitride nanosheets to absorb and decompose these genetic remnants present in sewage system wastewater before they are able to enter and infect other microorganisms.
For their study, the scientists focused on plasmid-encoded antibiotic-resistant genes (ARG) coding for New Delhi metallo-beta-lactamase 1 (NDM1), which is resistant to multiple drugs. When the treated nanosheets were combined in solution with the ARGs and exposed to ultraviolet light, they were 37 times better at killing the genetic remnants when compared to graphitic carbon nitride alone.
The study, which was performed under the guidance of the Rice-based Nanosystems Engineering Research Center for Nanotechnology-Enabled Water Treatment (NEWT), has been described in Environmental Science and Technology—a journal from the American Chemical Society.
This study addresses a growing concern, the emergence of multidrug resistant bacteria known as superbugs. They are projected to cause 10 million annual deaths by 2050. As an environmental engineer, I worry that some water infrastructure may harbor superbugs. For example, a wastewater treatment plant in Tianjin that we’ve studied is a breeding ground, discharging five NDM1-positive strains for each one coming in. The aeration tank is like a luxury hotel where all bacteria grow.
Pedro Alvarez, Environmental Engineer, Rice University
Alvarez is also the director of the NEWT Center.
Alvarez continued, “Unfortunately, some superbugs resist chlorination, and resistant bacteria that die release extracellular ARGs that get stabilized by clay in receiving environments and transform indigenous bacteria, becoming resistome reservoirs. This underscores the need for technological innovation, to prevent the discharge of extracellular ARGs.
“In this paper, we discuss a trap-and-zap strategy to destroy extracellular ARGs. Our strategy is to use molecularly imprinted coatings that enhance selectivity and minimize interference by background organic compounds,” he added.
Molecular imprinting is just like creating a lock that attracts a key, similar to natural enzymes with binding sites that accommodate only the accurately shaped molecules. In this study, graphitic carbon nitride molecules serve as the photocatalysts, or lock, which are tailored to absorb and subsequently destroy NDM1.
To produce the catalyst, the scientists initially coated the edges of the nanosheet with an embedded guanine, methacrylic acid, and polymer.
“Guanine is the most readily oxidized DNA base,” added Alvarez. “The guanine is then washed with hydrochloric acid, which leaves behind its imprint. This serves as a selective adsorption site for environmental DNA (eDNA).”
According to Danning Zhang, a graduate student at Rice University and the study’s co-lead author, carbon nitride was selected for the base nanosheets because it is easily available. It is also nonmetallic and hence safer to use.
Alvarez observed that all catalysts can efficiently eliminate ARGs from distilled water, but they are not as effective as in secondary effluent—a product of sewage treatment plants, obtained after removing organic compounds and solids.
In secondary effluent, you have reactive oxygen species scavengers and other inhibitory compounds. This trap-and-zap strategy significantly enhances removal of the eDNA gene, clearly outperforming commercial photocatalysts.
Pedro Alvarez, Environmental Engineer, Rice University
The team wrote that traditional disinfection processes such as ultraviolet radiation and chlorination employed at wastewater treatment plants, are moderately effective in eliminating antibiotic-resistant microorganisms but comparatively ineffective at eliminating ARGs. The researchers are hoping that their new technique can be adapted on a large scale.
Zhang stated that the laboratory is yet to conduct elaborate tests on other ARGs. “Since guanine is a common constituent of DNA, and thus ARGs, this approach should also efficiently degrade other eARGs,” he added.
In spite of its remarkable initial success, there is scope to enhance the existing process.
We have not yet attempted to optimize the photocatalytic material or the treatment process. Our objective is to offer proof-of-concept that molecular imprinting can enhance the selectivity and efficacy of photocatalytic processes to target eARGs.
Danning Zhang, Study Co-Lead Author and Graduate Student, Rice University
Qingbin Yuan of Nanjing Tech University, China, is the study’s co-lead author. Other co-authors are graduate students Ruonan Sun and Hassan Javed from Rice University, and Gang Wu, an assistant professor of hematology from The University of Texas Health Science Center at Houston’s McGovern Medical School.
Pingfeng Yu is the study’s co-corresponding author and is also a postdoctoral researcher at Rice University. Alvarez is the George R. Brown Professor of Civil and Environmental Engineering and also a professor of chemistry and of chemical and biomolecular engineering.
The study was supported by the National Science Foundation and the National Natural Science Foundation of China.