Despite the advantages of mammalian cell display technology, a few attributes of displayed proteins like their re-entry mechanism into cells remain unclear. In an article recently published in the journal Analytical Chemistry, the authors present an overview of the displayed protein’s endocytosis mechanism, motility behavior, and transport kinetics with the help of HaloTag, which is used as the displayed protein and quantum dot-based single-particle tracking.

Study: Quantum Dots Tracking Endocytosis and Transport of Proteins Displayed by Mammalian Cells. Image Credit: Van Pympk/Shutterstock.com
Tracking Display Proteins
Mammalian cell display technology has the potential to lead to protein folding and facilitate various functions related to post-translational processing. Thus, the obtained products closely mimic the natural biological protein molecules from their molecular structure to biological functions. However, core issues in the reentry mechanism of display protein cells remain unclear through mammalian cell display technology, thus limiting its widespread application.
The dynamic processes in biological systems can be studied using single-particle tracking. It is a microscopy technique used to visualize an individual particle trajectory. The tracked biomolecules are fluorescently labeled, which allows their in situ tracking for a long time.
Due to the unique optical properties of quantum dots (QDs), they have advantages in fluorescence imaging. The detection sensitivity of QDs increased with the high fluorescence intensity, enabling contrast images. Moreover, QDs' photostability facilitates their long-time tracking.
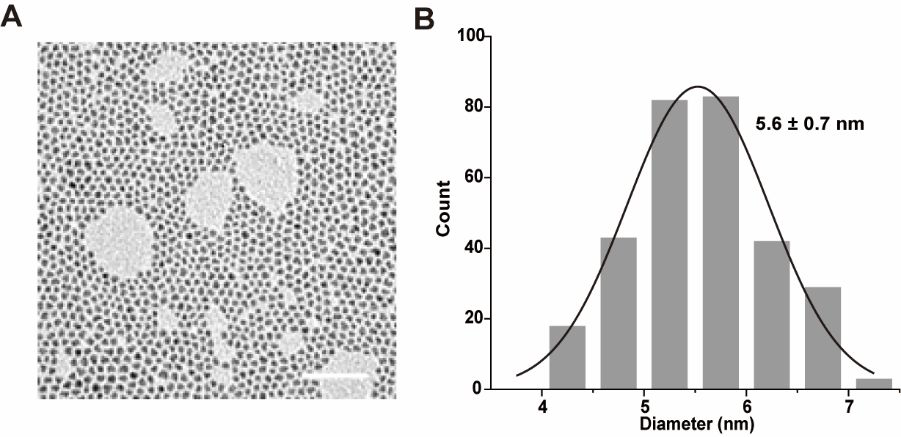
Transmission electron microscopy (A) and particle size statistics (B) of oilsoluble quantum dots. (Scale bar: 50 nm). Image Credit: Zhang, M Q et al., Analytical Chemistry
QDs Labelled HaloTags as Displayed Proteins
In the present work, the authors inserted HaloTag into the expression vector display and displayed it on the cell surface. The introduced HaloTag specifically binds to ligands that are halo derived via covalent bonding. Further, HaloTags were fluorescently labeled using QDs.
Employing single-particle tracking of the QDs, the authors found that HaloTag is not attached to the cell membrane but absorbed by the cell in a temperature- and time-dependent manner. The authors investigated the HaloTag endocytosis mechanism by combining the dominant negative mutation experiments with inhibitor-treated cells. The results revealed that entry of the HaloTag into the cells is through endocytosis mediated by clathrin.
The authors analyzed the QD-labeled HaloTag’s movement trajectories, and the results revealed that HaloTag transport occurs in three stages. Through colonization studies, the authors confirmed the aggregation of HaloTag in lysosomes. The real-time data demonstrated the transportation of HaloTag along the cytoskeleton.
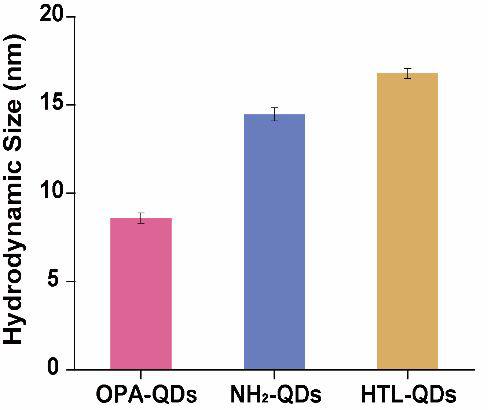
The hydrodynamic size was 8.6 ± 0.3 nm for OPA-QDs, 14.5 ± 0.4 nm for NH2-QDs, and 16.8 ± 0.3 nm for HTL-QDs. (One sample was repeated three times). Image Credit: Zhang, M Q et al., Analytical Chemistry
Research Findings
The authors performed hydrophobic encapsulation to obtain water-soluble QDs without disturbing the fluorescence property or functional groups on QDs. Thus, prepared QDs transferred into the aqueous phase without affecting the optical properties. The modification of QDs with polyethylene glycol (PEG) enhanced specificity in the adsorption of the materials and introduced the amino groups on the surface of QDs for subsequent reaction with the succinimidyl esters.
Transmission electron microscopy (TEM) results revealed that the hydrophobic QDs had an average particle size of 5.6 nanometers. Further results from dynamic light scattering (DLS) revealed that the water-soluble QDs (OPA-QDs) had a hydrodynamic size of 8.6 nanometers, and their modification with PEG and functionalization with HaloTag ligands increased their size to 16.8 nanometers.
The zeta potential of OPA-QDs, amino-modified (NH2) -QDs, and HaloTag ligand-functionalized quantum dots (HTL-QDs) was −38.7 millivolts, −17.1 millivolts, and −20.4 millivolts, respectively. This variation of zeta potential is because, in the first case, PEG increases the surface potential. In the second case, the reaction between the amino group and succinimide ester decreased the surface potential. Moreover, the electrophoretic mobility was in the order: HTL-QDs> NH2-QDs>OPA-QDs. The fluorescence results revealed that the binding of QDs to HaloTag-MDCK cells was via covalent bonds.
The incubation of HTL-QDs with the Martin-Darby canine kidney (MDCK) cells at 37 degrees Celsius for 0.5 hours revealed colonized signals of QDs and membrane dye (DiO) in the cytoplasm, suggesting the internalization of HaloTag by the cells. With the increase in incubation time, fluorescence from QDs intensified in the cytoplasm, and the DiO signal colocalized them, suggesting the entrapping of QDs-HaloTag inside nuclear endosomes. The authors confirmed that the QDs accumulated near the nucleus and did not always enter inside it.
To verify the clathrin-mediated endocytosis of displayed proteins, the authors observed the internalization of QDs-HaloTag after the overexpression of AP180C on cells, and the results revealed that the uptake of displayed HaloTag was blocked in the AP180C overexpressed cells.
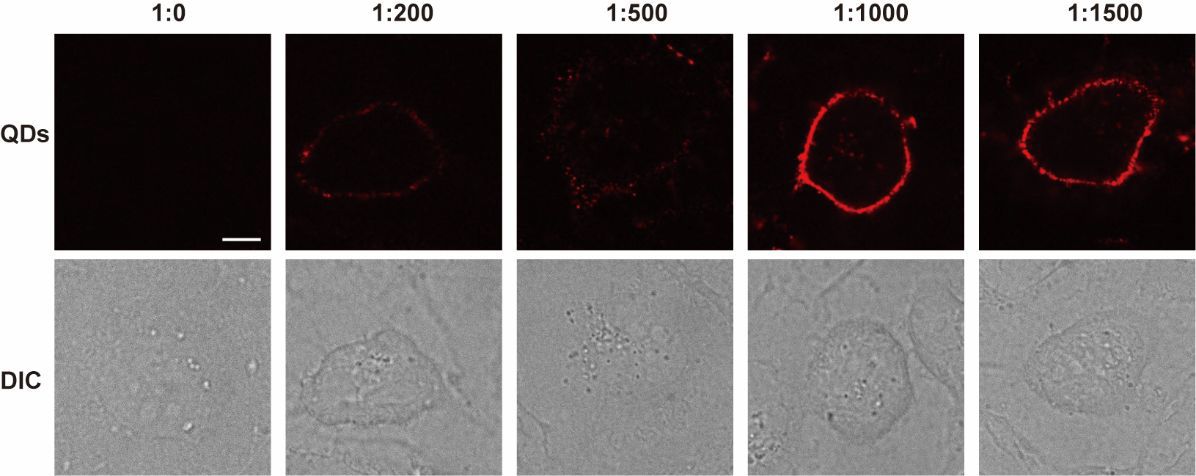
Incubation of MDCK cells with different ratios of HaloTag ligand-modified quantum dots. (Scale bar: 10 μm). Image Credit: Zhang, M Q et al., Analytical Chemistry
Conclusion
In this study, the authors labeled the HaloTag on the MDCK cell surface with HTL-QDs and studied the display protein’s endocytosis and post-endocytosis transport using living cells. The authors found the blockage in HaloTag uptake that inhibited clathrin function, suggesting the clarithin-mediated entry of HaloTag. The real-time studies revealed the movement of displayed proteins along the microtubules and microfilaments. The research findings revealed that intracellular processes like internalization and recycling determined the total amount of dynamic proteins on the cell surface.
Disclaimer: The views expressed here are those of the author expressed in their private capacity and do not necessarily represent the views of AZoM.com Limited T/A AZoNetwork the owner and operator of this website. This disclaimer forms part of the Terms and conditions of use of this website.
Source:
Zhang, M Q., Wang, Z G., Fu, D D., Zhang, J M., Liu, H Y., Liu, S L and Pang, D W. Quantum Dots Tracking Endocytosis and Transport of Proteins Displayed by Mammalian Cells (2022). Analytical Chemistry. https://pubs.acs.org/doi/10.1021/acs.analchem.2c00411