Reviewed by Alex SmithJun 17 2022
Detailed knowledge of nature’s smallest biological building elements — biomolecules — is essential to generating new medications and vaccinations. Researchers at the Chalmers University of Technology in Sweden have developed a revolutionary microscopy approach that allows proteins, DNA, and other small biological particles to be investigated in their natural conditions in an entirely new way.
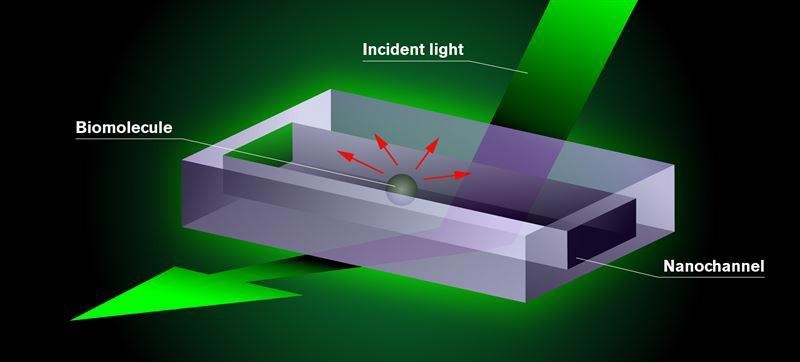 New microscopy method. The biomolecules the researchers want to study are placed in a chip consisting of tiny nano-sized tubes–nanochannels. Test fluid is added to the chip, which is mounted in an optical dark-field microscope and illuminated with visible light. The molecule appears as a dark shadow moving freely inside the channel on a screen connected to the microscope. The darkness of the shadow is proportional to the mass of the molecule. This is the result of an interference effect in the interaction of the light with the nanochannel and the molecule. Image Credit: Envue Technologies | Maja Saaranen.
New microscopy method. The biomolecules the researchers want to study are placed in a chip consisting of tiny nano-sized tubes–nanochannels. Test fluid is added to the chip, which is mounted in an optical dark-field microscope and illuminated with visible light. The molecule appears as a dark shadow moving freely inside the channel on a screen connected to the microscope. The darkness of the shadow is proportional to the mass of the molecule. This is the result of an interference effect in the interaction of the light with the nanochannel and the molecule. Image Credit: Envue Technologies | Maja Saaranen.
The development of medicines and vaccines takes a lot of time and money. It is critical, then, to be able to simplify the process by looking at how specific proteins, for example, behave and react with one another. Chalmers’ novel microscopy technology may allow for the identification of the most promising candidates at an earlier stage.
The method might potentially be used to study how cells communicate with one another by secreting chemicals and other biological nanoparticles. These mechanisms, for example, play a vital part in the immune response.
Revealing its Silhouette
Biomolecules are tiny and elusive, but they are essential as they are the building blocks of all living things. Researchers must now label them with a fluorescent label or connect them to a surface in order to get them to expose their secrets using optical microscopy.
With current methods you can never quite be sure that the labeling or the surface to which the molecule is attached does not affect the molecule’s properties. With the aid of our technology, which does not require anything like that, it shows its completely natural silhouette, or optical signature, which means that we can analyze the molecule just as it is.
Christoph Langhammer, Research Leader and Professor, Department of Physics, Chalmers University of Technology
Langhammer created the novel approach in collaboration with physics and biology researchers at Chalmers and the University of Gothenburg.
The novel microscopy approach involves flushing molecules or particles that the researchers seek to analyze via a chip with nanochannels, which are small nano-sized tubes. The chip is then filled with a test solution and irradiated with visible light. The interaction between the light, the molecule, and the microscopic fluid-filled channels cause the molecule within to appear as a dark shadow on the microscope’s screen, which can then be viewed.
Researchers may use it to not only view the biomolecule but also to calculate its mass and size, as well as acquire indirect information about its shape — something that was previously impossible to do with a single technique.
Acclaimed Innovation
Nanofluidic scattering microscopy, a novel technology, was recently published in the scientific journal Nature Methods. The Royal Swedish Academy of Engineering Sciences, which recognizes a number of research initiatives with the potential to transform the world and deliver real benefits every year, has also praised the achievements.
Through the start-up Envue Technologies, which won the “Game Changer” title at this year’s Venture Cup competition in Western Sweden, innovation has also made its way into society.
Our method makes the work more efficient, for example when you need to study the contents of a sample, but don’t know in advance what it contains and thus what needs to be marked.
Barbora Špačková, Researcher, Department of Physics, Chalmers University of Technology
Špačková is a Chalmers researcher who developed the theoretical foundation for the novel approach and then conducted the first practical investigation using the technology during her time there.
The researchers are now working to improve the nanochannel design in order to locate even smaller molecules and particles that are now undetectable.
The aim is to further hone our technique so that it can help to increase our basic understanding of how life works, and contribute to making the development of the next generation medicines more efficient.
Christoph Langhammer, Research Leader and Professor, Department of Physics, Chalmers University of Technology
How the Technique Works
The molecules or particles that the researchers seek to analyze are placed on a chip that has nanochannels, which are small tubes filled with test fluid. The chip is placed in a dark-field optical microscope and lit with visible light.
The molecule displays a dark shadow freely moving inside the nanochannel on the screen that depicts what may be viewed in the microscope. This is because the light is incident with both the channel and the biomolecule. The molecule’s optical signature is considerably enhanced as a result of the interference effect, which weakens the light precisely at the location where the molecule is present.
The amplification effect is stronger when the nanochannel is smaller, and the molecules visible are smaller. Biomolecules with molecular weights of roughly 60 kilodaltons and higher may now be analyzed using this approach. Larger biological particles, including extracellular vesicles and lipoproteins, and inorganic nanoparticles can be studied.
More About the Scientific Article and the Research
Barbora Špačková, Joachim Fritzsche, Daniel Midtvedt, Johan Tenghamn, Gustaf Sjösten, Henrik Klein Moberg, Hana Šípová-Jungová, Quentin Lubart, Daniel van Leeuwen, David Albinsson, Fredrik Westerlund, Elin K. Esbjörner, Giovanni Volpe, Mikael Käll, and Christoph Langhammer published the article in Nature Methods.
Chalmers University of Technology and the University of Gothenburg, both in Sweden, are where the researchers are active. Barbora Špačková is now forming her own research group at Prague’s Czech Academy of Sciences.
The Swedish Foundation for Strategic Research, as well as the Knut and Alice Wallenberg Foundation, financed the majority of the research. A portion of the study was carried out at the Chalmers Nanofabrication Laboratory, which is part of the Department of Microtechnology and Nanoscience (MC2) and is part of the Chalmers Excellence Initiative Nano.
Journal Reference:
Špačková, B., et al. (2022) Label-free nanofluidic scattering microscopy of size and mass of single diffusing molecules and nanoparticles. Nature Methods. doi.org/10.1038/s41592-022-01491-6.