Respiratory tract infections have been recognized to be one of the key causes of death worldwide. Several viral infections manifest similar symptoms, and clinically differentiating between them is challenging. However, proper diagnosis of these infections is extremely important to provide the best treatment to the patients.
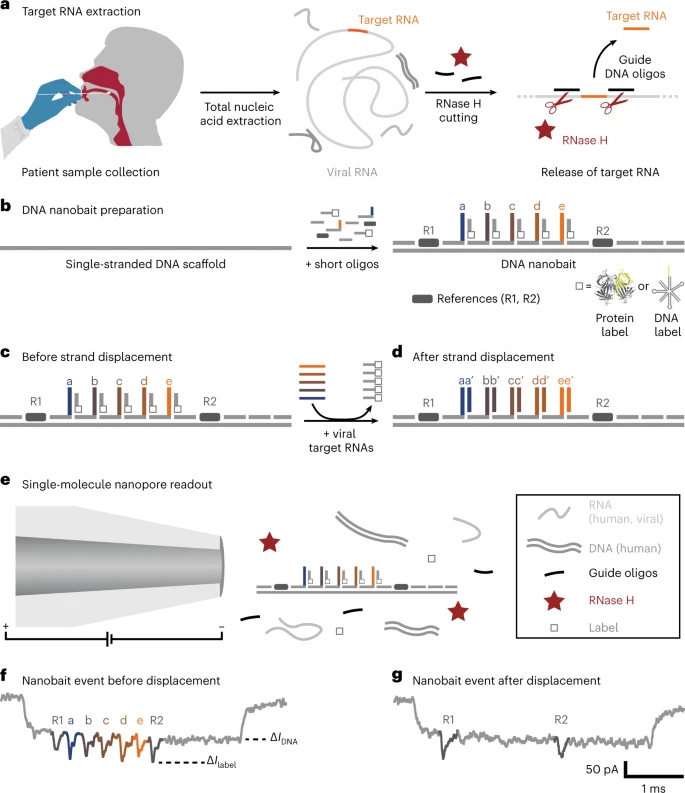
Self-assembled DNA nanobait strategy for multiplexed viral diagnostics. a, Oropharyngeal swab sample is collected from patients suspected to have COVID-19. The total nucleic acids were extracted (human and viral RNAs are shown in light grey; DNA is shown in dark grey), and the target RNA was cleaved out using programmable RNase H cutting. Such a treated sample was further tested for viral presence. b, Nanobait is made by mixing a single-stranded DNA scaffold (M13mp18 DNA) with short oligonucleotides where some of them carry complementary capture strands (a–e) for specific targets (a’–e’) in a target viral RNA. In addition, a partially complementary oligo with a structural label (protein or DNA based; white square) is added to each site to amplify the signal in nanopore recordings. We marked a sensing region with two references (dark grey). c,d, Nanobait before (c) and after (d) strand displacement reactions of five targets (coloured strands). If the targets are present, the five grey strands with labels are displaced. The two outer signals originate from reference structures that indicate the sensing region, and the five binding sites between the references are specific for the five different targets. e, Each nanobait is voltage driven through the nanopore and detected in a mixture of molecules. f, Typical nanopore current signature as a function of time for a nanobait as designed with five labels present. The first current drop corresponds to DNA (ΔIDNA) and the second to labels (ΔIlabel). g, Typical nanobait event after strand displacement of all the five targets. The presence of targets is detected by the missing downward signals specific to each target. Image Credit: Bošković, F. et al. (2023), Nature Nanotechnology
Recently, researchers at The University of Cambridge have focussed on developing an innovative method based on DNA nanobait, which can simultaneously detect multiple respiratory viruses and their variants in less than an hour. This study is available in Nature Nanotechnology.
Challenges in Respiratory Infection Diagnosis
Around 65% of viral infections are incorrectly diagnosed as pneumonia, and approximately 95% of misdiagnosed patients receive unnecessary antimicrobial treatments. The current coronavirus disease 2019 (COVID-19) pandemic, caused by the rapid outbreak of severe acute respiratory syndrome coronavirus-2 (SAR-CoV-2) virus, further underscores the diagnostic limitations of rapidly identifying the virus variants.
Due to genomic mutations, the continual emergence of SARS-CoV-2 variants has prolonged the pandemic despite the availability of COVID-19 vaccines. This virus has claimed millions of lives due to the lack of quick diagnostic measures. According to Professor Stephen Baker from the Cambridge Institute of Therapeutic Immunology and Infectious Disease, who co-authored the present study, “For patients, we know that rapid diagnosis improves their outcome, so being able to detect the infectious agent quickly could save their life,".
Some of the techniques used to identify viral variants are quantitative reverse transcription–polymerase chain reaction (qRT-PCR), followed by molecular sequencing. Although polymerase chain reaction (PCR)-based diagnostic methods are commonly used to identify nucleic acids from complex biological samples, they have limited multiplexing capabilities. Therefore, there is an urgent need for a novel diagnostic method to detect viruses as well as their variants in reduced sample volume.
Recently, nanotechnology techniques, such as nanopore sensing, have been applied for virus detection. This technique can differentiate between several nucleic acid species based on the uniquely engineered DNA nanostructure. These DNA nanostructures have the potential for multiplexed biosensing.
Development and Working Principle of DNA Nanobait
Scientists aimed to overcome the aforementioned diagnostic limitations using a nanotechnology-based approach. This method is based on a DNA nanobait, which can simultaneously detect multiple respiratory viruses and their variants. The detection method is based on obtaining swab samples of patients, followed by nucleic acid extraction and programmable RNase H cutting of viral RNA.
In this technique, the RNA targets are predominantly selected via oligonucleotides, a single-stranded DNA, which can bind at a specific region (both upstream and downstream) of the viral genome. Subsequently, RNase H was employed to digest the RNA sequence in RNA: DNA hybrids, and release the middle target RNA, which was identified using sequence-specific binding to the nanobait.
The genetically engineered DNA nanobait contained five binding sites, which could bind to as many RNA targets. The nanobait was assembled by combing a single-stranded DNA scaffold with several short complementary oligonucleotides. One end of the nanobait contained a sensing region, a DNA overhang complementary to the target sequence. A proper nanobait assembly was confirmed by electrophoretic mobility shift assay and atomic force microscopy (AFM) imaging tools.
A target sequence binds to one of six unpaired bases by replacing the oligonucleotide with the label at its complementary overhang. This method is known as toehold-mediated strand displacement. Therefore, the presence of the target nucleotide was detected by the absence of a label at the respective site.
A nanopore DNA sensor detects the nanobait based on its voltage-driven translocation. The negatively charged nanobait travels, through a narrow opening, towards a positively charged electrode in an electrolyte solution, eliciting a distinct current blockage signature. Each nanobait–nanopore event reveals the presence of RNA targets. The unique nanobait designs enabled the identification of targets originating from various parts of the same virus or numerous viral genomes.
Simultaneous Identification of Multiple Viral Variants
The newly developed nanobait can help identify multiple respiratory viruses, such as a respiratory syncytial virus (RSV), SARS-CoV-2, parainfluenza, and influenza. The authors demonstrated that the newly designed DNA nanobait could detect RSV based on site-specific displacement.
The induction of spikes after nanopore events indicated the absence of targets, which confirmed the presence of specific targets for RSV, SARS-CoV-2, rhinovirus, parainfluenza, and influenza. Importantly, the five sites of nanobait enabled simultaneous identification of wild-type SARS-CoV-2 and its four variants, namely, B.1, B.1.1.7 (Alpha), B.1.351 (Beta), and B.1.617 (Delta).
The DNA nanobait was programmed using a single-nucleotide mismatch, which enabled the identification of the SARS-CoV-2 original strain and newly emerged variants. Notably, in a nanopore sensor, dark-colored bars were observed for the original SARS-CoV-2 strain and light-colored bars for all four variants.
The developmental procedure of the DNA nanobaits is extremely advantageous because it skips preamplification and purification steps, which are potentially time-intensive and expensive steps.
While discussing the future application of the nanobait, Professor Ulrich Keyser of Cavendish Laboratory, who also co-authored the study, said, "Nanobait is based on DNA nanotechnology and will allow for many more exciting applications in the future," He further added, "For commercial applications and roll-out to the public we will have to convert our nanopore platform into a hand-held device."
Reference
Bošković, F. et al. (2023) Simultaneous identification of viruses and viral variants with programmable DNA nanobait. Nature Nanotechnology. https://doi.org/10.1038/s41565-022-01287-x.
Disclaimer: The views expressed here are those of the author expressed in their private capacity and do not necessarily represent the views of AZoM.com Limited T/A AZoNetwork the owner and operator of this website. This disclaimer forms part of the Terms and conditions of use of this website.