May 16 2008
Using two sophisticated imaging techniques, scientists at the U.S. Department of Energy's Brookhaven National Laboratory and collaborators at Stony Brook University, University College London, and Washington University School of Medicine have captured molecular-level "snapshots" of the cellular machinery that assembles hairlike projections on certain bacterial cells. These projections, called pili, enable the bacteria to attach to and infect bladder and kidney cells.
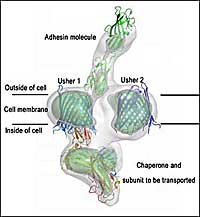 How bacteria grow their surface pili (fibers). This picture shows a cryo-EM structure (in gray contour) of the pilus assembly apparatus with the pilus tip molecule - the adhesin - making its way out of the cell. The crystals structures of individual proteins are placed into the EM structure. The cell membrane-embedded usher structure is determined in this work.
How bacteria grow their surface pili (fibers). This picture shows a cryo-EM structure (in gray contour) of the pilus assembly apparatus with the pilus tip molecule - the adhesin - making its way out of the cell. The crystals structures of individual proteins are placed into the EM structure. The cell membrane-embedded usher structure is determined in this work.
In a paper appearing in the May 16, 2008, issue of the journal Cell, the team describes how their newly revealed structures provide a mechanistic understanding of the pilus assembly process - which could lead to new ways to thwart or treat urinary tract infections.
The research focuses on the surface pilus of E. coli bacteria, which cause urinary tract infections in 1 in 10 women in the U.S. every year. E. coli normally reside in the human gut, where they are harmless. But if the microbes get into the urinary tract, they produce sticky tendrils known as pili. These pili allow the bacteria to recognize, stick to, and invade cells lining the bladder and kidneys, and go on to cause infections.
"Understanding how the bacteria assemble these pili is a first step toward finding an antimicrobial inhibitor that interferes with the process," said Brookhaven biophysicist and study co-author Huilin Li.
According to Li, there have been extensive studies on many aspects of E. coli pili biogenesis. "The basic process and major molecular players - including the structures of many pilus components and the fully assembled hairlike projections - are known. What has been unknown is this difficult albeit crucial question: how the individual components are assembled and moved across the bacterial outer membrane," Li said. The new images help to answer this question.
A protein known as usher, which traverses the cell membrane, is the engine of the pilus assembly line. The usher catalyzes the assembly of individual components into a pilus and provides a pathway for the assembled pilus to move out of the cell and extend from the cell surface. The Cell paper describes a new high-resolution image of the translocation domain of the usher protein, as well as a broader view of the assembly apparatus caught in action with the pilus tip making its way out of the cell.
The usher protein structure was determined by Han Remaut, University College London, using x-ray crystallography at the European Synchrotron Radiation Facility in Grenoble, France, under the guidance of Gabriel Waksman, University College London, and Scott Hultgren, Washington University, St. Louis. In this technique, scientists analyze how a beam of x-rays bounces off or is scattered by a molecule to determine the relative positions of its individual atoms.
The images revealed a protein "dimer" made of two identical ushers, each forming a barrel-shaped pore with a "plug" blocking the opening of the barrel. The plug is suspected to move away, making space for the pilus to move through the membrane as it is formed.
The structure of a partially formed pilus in association with the usher dimer was determined by single-particle cryo-electron microscopy (cryo-EM) by postdoctoral researchers Chunyan Tang and Tao Wang working with Li at Brookhaven Lab. The structure of this complex, which had been isolated by microbiologist David Thanassi and colleagues at Stony Brook University, confirmed the dimeric form of the functional usher protein, supporting the crystallography result as well as previous research on the structure by Li's group.
Interestingly, says Li, the pilus tip fiber appears to grow from only one of the two usher pores. However, both ushers are involved in recruiting and incorporating the pilus components into the growing pilus.
"Both techniques were essential to gaining insights into the pilus formation process," said Li. "Crystallography is good at revealing very high resolution structures of relatively small proteins or individual proteins, but not so good at imaging multiple protein assemblies. In contrast, cryo-EM is good at revealing the structure of large protein assemblies, but at lower resolution. When combined, the high-resolution structures of individual pieces (or component proteins) from crystallography can be placed into the cryo-EM map of the whole assembly, yielding a comprehensive understanding of the underlying biology," he explained.
The scientists still do not know how the assembly is initiated or how the plugged pore is opened for secretion. Future research will address these questions, as well as whether the molecular mechanism observed here operates in other related systems.
This research was funded by the National Institute of General Medical Sciences and National Institute of Allergy and Infectious Diseases, both of the National Institutes of Health, and by Brookhaven's Laboratory Directed Research and Development Program. The research team also included Nadine Henderson, Stony Brook University, and Jerome Pinkner, Washington University.