In this interview, AZoNano speaks with Associate Professor Ebbe Sloth Andersen from Aarhus University about a new software-based 'RNA origami' technique that can be used to design RNA molecules that fold into compact and rigid shapes. We discuss the applications of RNA nanotechnology, the different structures that can be designed with this technique, and what this research might mean for the future of RNA-medicine.
Please could you introduce yourself and your current research activities at the Andersen Lab for Biomolecular Design?
I am an Associate Professor at the Interdisciplinary Nanoscience Center (iNANO) at Aarhus University, Denmark. In my research group, we are working on mastering the design of DNA, RNA and protein nanostructures, testing our abilities by using advanced characterization techniques such as atomic force microscopy (AFM) and cryo-electron microscopy (cryo-EM). We further use the designed molecular structures to demonstrate novel applications in medicine and synthetic biology.
"Nanosatellite explores folding space". Artist rendering by Cody Geary showing an RNA satellite exploring the possibilities for folding and applications in nanomedicine and synthetic biology.
How did you become involved in this field of research?
I initially researched RNA structures of HIV, but got fascinated by the possibilities of designing biomolecules when reading a paper on DNA origami back in 2006. Luckily, I had the chance to join a center for DNA nanotechnology that allowed me to develop software and implement DNA origami designs experimentally. When I started my own research group later on, I gradually gravitated back to RNA by developing the RNA origami method and investigating its applications.
Over the past few years, particularly due to the COVID-19 pandemic, RNA biology has been brought firmly into the public sphere. How would you define RNA nanotechnology?
RNA biology and nanotechnology have indeed entered the spotlight with the success of mRNA vaccines, where mRNA design and nanoparticle formulation have played central roles. The approach that we call RNA nanotechnology is concerned with designing the shape and function of the RNA particles to provide additional benefits to RNA medicine.
Could you discuss the newly developed ‘RNA origami’ technique and how it is used to create RNA nanostructures?
The RNA origami technique is a set of software tools that allows you to design RNA molecules that fold into compact and rigid shapes. The process starts by constructing a blueprint using a set of compatible RNA modules. The software can show you the atomic model of your blueprint, and when you are happy with the result, it can provide you with a sequence to order from a gene synthesis company that will fold into your desired shape.
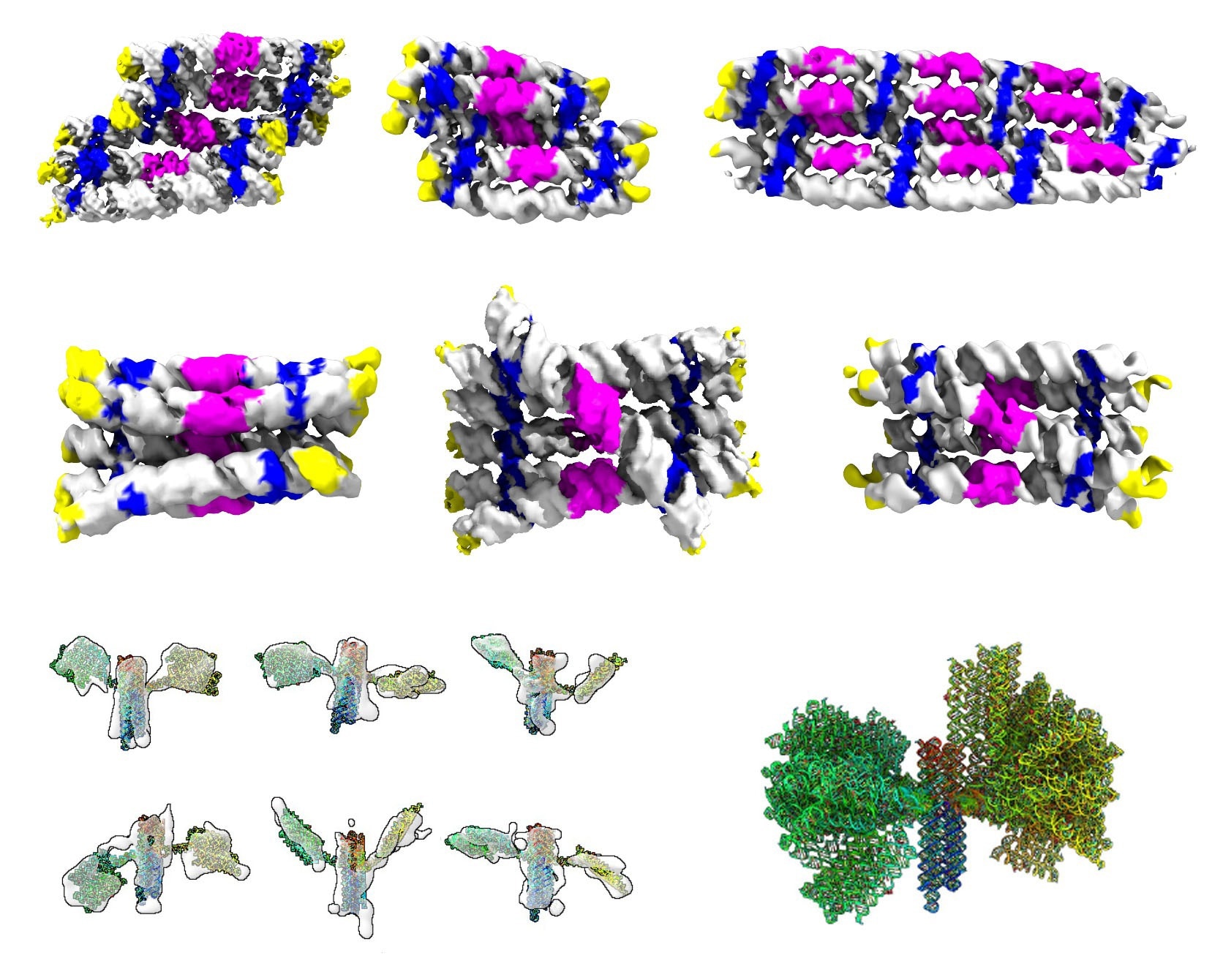
Gallery of the different RNA origami structures that were determined by cryogenic electron microscopy and tomography. Top rows show structures of RNA rectangles and cylinders colored by RNA motifs. Bottom row shows structures of the nanosatellite colored in rainbow in the reading direction of an RNA strand. © Ebbe Sloth Andersen, PhD
This technique can be used to design RNA nanostructures that self-assemble through cotranscriptional folding. What is the relationship between cotranscriptional folding and gene regulation?
A benefit of RNA origami architecture is that it is compatible with cotranscriptional folding, which means that the RNA will fold immediately as it is synthesized by an RNA polymerase. This means that you can produce it on a large scale without further preparation steps and that it can be genetically encoded and fold directly in cells. Cotranscriptional folding can lead to misfolds and kinetic traps and is known to result in riboswitches that are involved in gene regulation in cells.
Why are RNA folding mechanisms so exciting for medical research, and more specifically, nanomedicines?
RNA folding mechanisms are used by biology to assemble complex RNA machines like the ribosome and regulate gene expression. The assembly properties can be used to create compact and stable RNA particles for efficient delivery.
The regulation mechanisms can be used in RNA medicine to control the properties and conditional activation of the RNA to be delivered. With these RNA mechanisms built into mRNA vaccines, it may be possible to restrict protein expression to only very specific cells.
What are some of the RNA bundle and sheet designs that the team designed?
In the paper, we designed RNA sheets and bundles and finally combined them in the design of a satellite shape inspired by the Hubble space telescope. The shapes we designed are not fit for practical use but were used to challenge our design software and push the boundaries of the possible.
Doing this exploratory research paid back by providing us with several important insights on improving our design process, revealing a surprising “folding trap” and highlighting flexible features of RNA molecules in general.
What were the benefits of utilizing Cryo-Electron Microscopy and Small Angle X-Ray Scattering in this investigation?
We have earlier used atomic force microscopy (AFM) for our characterization of RNA origami structures, but with this method, the RNA is deposited on a surface, which might influence the result. Cryo-EM and SAXS are solution methods where we can study the RNA molecules in their free-floating native state. Furthermore, cryo-EM is proven to be an efficient method for obtaining high-resolution structural information of RNA structures and also be able to study the flexibility of RNA in solution.
SAXS provides a nice complementary method to look at larger conformational changes over time.

© PolakPhoto/Shutterstock.com
What are the next steps for this investigation? Are there any directions the group is working towards?
The current study is just the beginning of an exploration of RNA design and folding. With the cryo-EM pipeline, we can now get high-quality feedback on our design efforts. We will in the future work on making bigger and more advanced designs. We are currently working on chemical modification of our RNA structures and on expressing RNA nanostructures in cells.
Where can our readers go to stay up to date with this research and your lab’s activities?
About Ebbe Sloth Andersen, Ph.D.
 Ebbe Sloth Andersen is an Associate Professor at the Interdisciplinary Nanoscience Center at Aarhus University, Denmark, and is also affiliated with the Department of Molecular Biology and Genetics. He did his Ph.D. studies on HIV RNA structure and function in the laboratory of Professor Jørgen Kjems at Aarhus Univeristy and his Postdoc work on DNA origami in the context of the Centre for DNA Nanotechnology lead by Professor Kurt Gothelf.
Ebbe Sloth Andersen is an Associate Professor at the Interdisciplinary Nanoscience Center at Aarhus University, Denmark, and is also affiliated with the Department of Molecular Biology and Genetics. He did his Ph.D. studies on HIV RNA structure and function in the laboratory of Professor Jørgen Kjems at Aarhus Univeristy and his Postdoc work on DNA origami in the context of the Centre for DNA Nanotechnology lead by Professor Kurt Gothelf.
As Assistant Professor he has been a visiting associate at California Institute of Technology working with Professor Paul Rothemund, inventor of the DNA origami method, to develop the RNA origami methodology. Ebbe has been running his own research group since 2012 with a focus on designing DNA and RNA nanoscale devices for applications in medicine and synthetic biology.
Disclaimer: The views expressed here are those of the interviewee and do not necessarily represent the views of AZoM.com Limited (T/A) AZoNetwork, the owner and operator of this website. This disclaimer forms part of the Terms and Conditions of use of this website.