Nov 19 2009
Chromosomal DNA is normally coiled and compacted via interactions with histone proteins to form material called chromatin. Chemical modification of the histones can profoundly alter how the chromatin is arranged, and this represents an important mechanism by which cells regulate gene expression.
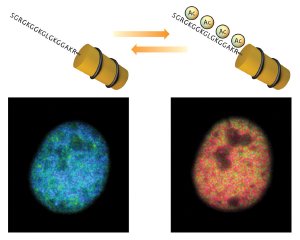 Tracking changes in histone acetylation. Histac produces a strong FRET signal (blue) when chromatin is unacetylated (left), but this signal diminishes when the chromatin is acetylated (right).
Tracking changes in histone acetylation. Histac produces a strong FRET signal (blue) when chromatin is unacetylated (left), but this signal diminishes when the chromatin is acetylated (right).
One example is the addition of acetyl groups by histone acetyltransferase (HAT) enzymes. HATs mark histone H4 at multiple sites, and these acetylations are subsequently recognized by regulatory proteins of chromatin such as BRDT. However, this is a highly dynamic and reversible process, and these acetylations are routinely removed by histone deacetylase enzymes (HDACs).
To develop tools for tracking histone acetylation in live cells, Minoru Yoshida of the RIKEN Advanced Science Institute in Wako paired his knowledge of histone acetylation with colleague Kazuki Sasaki’s expertise in fluorescent biosensor design. The result was Histac—a highly sensitive genetically encoded indicator that enables real-time imaging of changes in H4 acetylation1.
Their sensor is based on Förster resonance energy transfer (FRET), which makes use of two distinct ‘donor’ and ‘acceptor’ fluorescent proteins. When the donor is excited by light, it releases energy that excites the acceptor and causes it to produce fluorescent signal, but only when the two are close together. In Histac, they are linked by H4 and a chunk of BRDT; they can interact in the unacetylated state, but when the H4 segment is acetylated, subsequent interaction with BRDT separates the fluorescent proteins and greatly decreases the FRET signal (Fig. 1).
Yoshida and colleagues demonstrated that Histac effectively incorporates into cellular chromatin, and subsequent treatment with HDAC inhibitor trichostatin A yielded a strong signal change, indicating successful detection of elevated histone acetylation. Subsequent experiments enabled Yoshida and Sasaki to directly observe histone deacetylation as cells undergo mitosis, followed by an increase in acetylation once cell division was complete—a somewhat surprising finding. “This dramatic decrease in histone H4 acetylation in mitosis was a matter of dispute,” says Yoshida. Interestingly, they were even able to observe subtle variations in the rate of histone deacetylation at different regions within a single cell nucleus.
Having demonstrated its effectiveness and sensitivity, Yoshida and colleagues are expanding their work with Histac and developing additional tools that will enable them to analyze complex patterns of chromatin modification relating to embryonic development. “It has been proposed that epigenetic changes—including histone acetylation—are important for embryonic reprogramming and tissue differentiation,” says Yoshida. “We therefore would like to visualize histone acetylation in embryonic cells under various conditions.”
Sasaki, K., Ito, T., Nishino, N., Khochbin, S. & Yoshida, M. Real-time imaging of histone H4 hyperacetylation in living cells. Proceedings of the National Academy of Sciences USA 106, 16257–16262 (2009). article