Nov 25 2009
With a new screening method, based on a DNA-encoded substance collection, ETH researchers from the a group headed by Dario Neri, professor of bio-macromolecules at the Institute for Pharmaceutical Sciences, are attempting to revolutionise the search for potentially active substances for medicines. The method is simple, fast and cheap. In the context of this project the group is already searching for new active substances against rheumatoid arthritis.
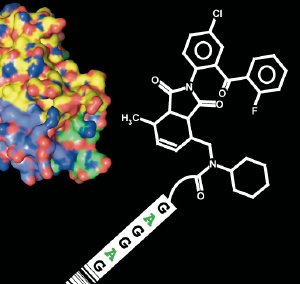 Researchers search through thousands of molecules for potential active substance candidates which bind to a target protein (left). The molecules can be identified on the basis of a small strand of DNA. (Image: Fabian Buller/ETH Zurich)
Researchers search through thousands of molecules for potential active substance candidates which bind to a target protein (left). The molecules can be identified on the basis of a small strand of DNA. (Image: Fabian Buller/ETH Zurich)
Dario Neri, Professor at the Institute for Pharmaceutical Sciences of the ETH Zurich, speaks of a possible revolution in the screening for active substances, which is greatly facilitated by the use of “DNA-encoded chemical libraries”. These involve a collection of innumerable different small molecules, to which a short strand of DNA has been attached as an individual identity tag, similar to a bar code on a product.
The new method, in which Neri's graduate student Fabian Buller is heavily involved, and which is financially supported by the agency for the promotion of innovation KTI and the Gebert Rüf foundation, could also be of interest to the pharmaceutical industry: the reason for this is that procedures for testing possible active substances are so far time-consuming and very expensive. As soon as a medically relevant target protein is discovered, substance libraries, containing about one million substances, are tested in the pharmaceutical industry with robotic systems. Furthermore, the follow-up of the “hits” is often so expensive that essentially only large companies can use this method.
Simpler, faster, cheaper
The method which was published by Fabian Buller and other scientists in the journal “Chemistry & Biology”, simplifies and miniaturises the active substance screening. DNA-encoded substance libraries can contain millions and even billions of different substances and can be placed in so-called Eppendorf vessels, a miniature reaction vessel holding, for example, half a milliliter. Thus, for the first time, a large number of small molecules can be tested rapidly and economically on potential drug targets. The small organic binders are present in the library only in extremely small concentrations, so that even the smallest quantities of target protein are sufficient to perform a screening of the entire collection of substances.
In order to find potentially active substances, the researchers mix their substance library with an immobilised target protein in a single experiment. Molecules which do not bind to it can be washed away, the “hits”, however, can be rapidly identified through the DNA code using a polymerase chain reaction (PCR), which multiplies the DNA, and subsequent high-throughput sequencing.
Hit from 4000 substances
In order to test the method, Buller first prepared a collection of 4000 small molecules. These were examined for interactions with tumor necrosis factor (TNF). TNF is a signal protein of the immune system, which is formed during inflammation, for example in rheumatoid arthritis. There are several drugs on the market for this disease which act on TNF, and sell for billions. These drugs are, however, based exclusively on antibodies and not on small organic molecules.
From the 4000 substances the researchers were able to find a number of hits which interacted with the TNF-protein complex. After chemical refinement they went on to test the best candidate in a cell culture experiment. The substance they discovered allowed the cells to survive, by blocking the TNF signal. It could be thus shown for the first time that it should be possible to develop a TNF blocker based on a small molecule.
Next generation of TNF blockers
Current medicines against TNF based on antibodies can have certain disadvantages: They could be immunogenic and thus trigger an immune response in the body after repeated use. In this case they can only be administered for a short time forcing the patients to change to a different medicine. Antibodies must also be injected with a syringe. The advantage of a TNF blocker based on a small molecule would be that it is non-immunogenic and that it would be more convenient for the patient, as the medicine could be administered orally.
The molecules “fished” with the new screening method from a sea of substances are still not finished active substances and must be improved and tested clinically before they can be used in patients. This example shows, however, that the comparatively simple new screening method rapidly supplies promising results for a “hit”.
Moreover, the substance collection can be rapidly expanded. The ETH researchers have already extended their DNA-encoded molecules library to one million substances. The Neri group wants to develop this collection further to more than 100 million different small molecules in the future. In this way, scientists want to optimize active substance screening and develop small molecules against clinically relevant target proteins quickly and economically - and thus also find new drugs to combat rheumatoid arthritis.