Oct 5 2010
The enigmatic Möbius strip has long been an object of fascination, appearing in numerous works of art, most famously a woodcut by the Dutchman M.C. Escher, in which a tribe of ants traverses the form's single, never-ending surface.
Scientists at the Biodesign Institute at Arizona State University's and Department of Chemistry and Biochemistry, led by Hao Yan and Yan Liu, have now reproduced the shape on a remarkably tiny scale, joining up braid-like segments of DNA to create Möbius structures measuring just 50 nanometers across—roughly the width of a virus particle.
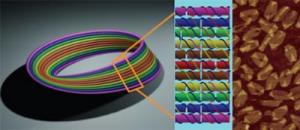 This is the design for the DNA Möbius strip. Single-stranded viral DNA is used as scaffolding and 164 short segments of DNA are used as staple strands, to create the nanostructure.
This is the design for the DNA Möbius strip. Single-stranded viral DNA is used as scaffolding and 164 short segments of DNA are used as staple strands, to create the nanostructure.
Eventually, researchers hope to capitalize on the unique material properties of such nano-architectures, applying them to the development of biological and chemical sensing devices, nanolithography, drug delivery mechanisms pared down to the molecular scale and a new breed of nanoelectronics.
The team used a versatile construction method known as DNA origami and in a dramatic extension of the technique, (which they refer to as DNA Kirigami), they cut the resulting Möbius shapes along their length to produce twisted ring structures and interlocking loops known as catenanes.
Their work appears in today's advanced online issue of the journal Nature Nanotechnology. Graduate students involved in this work include Dongran Han and Suchetan Pal in the Yan group.
Making a Möbius strip in the everyday world is easy. Cut a narrow strip of paper, bring the two ends of the strip close to each other so that they match, but give them a half-twist before fastening the ends together with a piece of scotch tape. The resulting Möbius strip, which has only one surface and one boundary edge, is an example of a topological form.
"As nanoarchitects," Yan says, "we strive to create two classes of structure—geometric and topological." Geometric structures in two and three dimensions abound in the natural world, from complex crystal shapes to starfish, and unicellular organisms like diatoms. Yan cites such natural forms as a boundless source of inspiration for human-designed nanostructures.
Topology, a branch of mathematics, describes the spatial properties of shapes that may be twisted, stretched or otherwise deformed to yield new shapes. Such shape deformations may profoundly alter the geometry of an object, as when a donut shape is pinched and stretched into a figure eight, but the surface topology of such forms is unaffected.
Nature is also rich in topological structures, Yan notes, including the elegant Möbius. The circulations of earth's warmer and cooler ocean currents for example, describe a Möbius shape. Other topological structures are common to biological systems, particularly in the case of DNA, the 3 billion chemical bases of which are packed by the chromosome inside the cell, using topological structures. "In bacteria, plasmid DNA is wound into a supercoil," Yan explains. "Then the enzymes can come in and cut and reconfigure the topology to relieve the torsion in the supercoil so that all the other cellular machinery can have access to the gene for replication, transcription and so forth."
To form the Möbius strip in the current study, the group relied on properties of self-assembly inherent in DNA. A strand of DNA is formed from combinations of 4 nucleotide bases, adenine (A), thymine (T), cytosine (C) and guanine (G), which follow one another on the strand like necklace beads. These nucleotide beads can bind to each other according to a strict rule: A always pairs with T, C with G. Thus, a second, complementary strand of DNA binds with the first to form the DNA double helix.
In 2006, Paul Rothemund at Cal Tech demonstrated that the process of DNA self-assembly could be used to produce pre-designed 2D nanoarchitectures of astonishing variety. Thus, DNA origami emerged as a powerful tool for nanostructure design. The method relies on a long, single stranded segment of DNA, used as a structural scaffold and guided through base pairing to assume a desired shape. Short, chemically synthesized "staple strands," composed of complementary bases are used to hold the structure in place.
After synthesis and mixing of DNA staples and scaffold strands, the structure is able to self-assemble in a single step. The technique has been used to produce remarkable nanostructures of smiley faces, squares, disks, geographic maps, and even words, at a scale of 100 nm or less. But the creation of topological forms capable of reconfiguration, like those produced by nature, has proven more challenging.
Once the tiny Möbius structures had been created, they were examined with atomic force- and transmission electron microscopy. The startling images confirm that the DNA origami process efficiently produced Escher-like Möbius strips measuring less than a thousandth the width of a human hair. Yan notes that the Möbius forms displayed both right and left handed twists. Imaging permitted the handedness or chirality of each flattened nanostructure to be determined, based on the height differences observed at the overlapping areas.
Next, the team demonstrated the topological flexibility of the Möbius forms produced, using a folding and cutting—or DNA Kirigami—technique. The Möbius can be modified by cutting along the length of the strip at different locations. Cutting a Möbius along its centerline yields a new structure—a looped form containing a twist of 720 degrees or 4 half-twists. The design, which the group calls a Kirigami-Ring is no longer a Möbius as it has two edges and two surfaces. The Möbius may also be cut along its length one-third of the way into its width, producing a Kirigami-Catenane—a Möbius strip interlinked with a supercoiled ring.
To accurately cut the Möbius nanostructures, a technique known as strand displacement was used, in which the DNA staples holding the central helix in place are outfitted with so-called toe-hold strands which protrude from the central helix. A complementary strand binds to the toehold segment, removing the staples and allowing the Möbius to fall open into either the Kirigami-Ring or Kirigami-Catenane.
Again, the successful synthesis of these forms was confirmed through microscopy, with the Kirigami-Ring structures gradually relaxing into figure eights.
Yan stresses that the success of the new study relied heavily on lead author Dongran Han's remarkable sense of three-dimensional space, allowing him to design geometrical and topological structures in his head. "Han and also Pal are particularly brilliant students," Yan says, pointing out that the complex conceptualization of the nanoarchitectures in their research is primarily performed without computer aid. The group hopes in the future to create software capable of simplifying the process.
"We want to push the Origami-Kirigami technology to create more sophisticated structures to demonstrate that we can make any arbitrary shape or topology using self-assembly," Han says.
Having made inroads into sculpture, painting and even literature, (particularly, the novels of French author Alain Robbe-Grillet), topological structures are now poised to influence scientific developments at the tiniest scale.
Source: http://www.asu.edu/