Oct 13 2010
Using large-scale computer simulations, researchers at the Georgia Institute of Technology have identified the most important factors affecting how molecules move through the crowded environment inside living cells. The findings suggest that perturbations caused by hydrodynamic interactions – similar to what happens when the wake from a large boat affects smaller boats on a lake – may be the most important factor in this intracellular diffusion.
A detailed understanding of the interactions inside cells – where macromolecules can occupy as much as 40 percent of the available space – could provide important information to the developers of therapeutic drugs and lead to a better understanding of how disease states develop. Ultimately, researchers hope to have a complete simulation of these cellular processes to help them understand a range of biological issues, from metabolism to cell division.
Sponsored by the National Institutes of Health, the research was reported Oct. 11 in the early online edition of the journal Proceedings of the National Academy of Sciences.
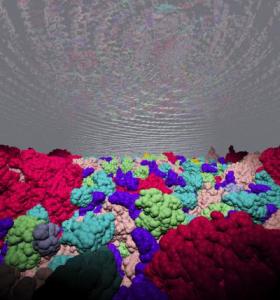 Cells are crowded with macromolecules such as proteins, RNA and DNA. Using a Brownian dynamics simulation of an Escherichia coli cytoplasm model, Georgia Tech researchers propose that crowding and hydrodynamic interactions dominate in vivo macromolecular motion.
Cells are crowded with macromolecules such as proteins, RNA and DNA. Using a Brownian dynamics simulation of an Escherichia coli cytoplasm model, Georgia Tech researchers propose that crowding and hydrodynamic interactions dominate in vivo macromolecular motion.
"We found that hydrodynamics – perturbation of the solvent with eddies and wakes created by molecules in this crowded environment – may be the dominant effect in intermolecular dynamics within cells," said Jeffrey Skolnick, director of the Center for the Study of Systems Biology at Georgia Tech. "The correlations created between molecules through this process have a lot of functional consequences for how collections of these molecules interact."
The motion of macromolecules within cells is normally random, occurring through Brownian motion that causes the molecules to diffuse through the cellular cytoplasm, which has viscosity similar to that of water. Researchers have studied the movement of fluorescent protein molecules injected into E. coli cells, but don't yet understand the forces affecting that motion. However, the measurements show that the fluorescent molecules move about 15 times more slowly inside the cell than they do in a test tube.
Using simulations that allowed them to adjust the impacts of natural forces, Skolnick and collaborator Tadashi Ando analyzed the activity of 15 different molecules in a portion – just one one-thousandth – of an E. coli cell. By altering those simulated forces in the computer, they attempted to determine what may cause the reduction in diffusion speed.
The most logical reason for that slowed movement is the crowded nature of cells, but Skolnick and Ando found that bumping into other molecules accounted for only a portion of the reduced molecular diffusion.
"If you are in a crowded room and want to walk to the bar, the other people slow you down," explained Skolnick, who is Georgia Research Alliance eminent scholar in computational systems biology. "In biological processes, if there are a lot of large molecules in the way, these protein molecules can't move as quickly. But our model showed that this crowding accounted for only about a third of the reduction measured experimentally."
The researchers also studied the hydrodynamic forces exerted by molecules on one another. These forces are comparable to the way in which the wake of a large boat on a lake affects smaller boats, or how a swimming whale might effect a school of small fish. The interaction causes correlated motion, which was known to be important in the movement of polymers and colloids studied earlier by chemists.
By turning off the other forces at work in their silicon world, the Georgia Tech researchers found that this correlated motion accounted for much more of the diffusion reduction than did the crowding.
"The hydrodynamic interactions create cooperative motion between the molecules," Skolnick explained. "We see long-lived correlations between the molecules, independent of size, in space and time. This suggests that these correlated motions may be extremely important in the dynamics of molecules."
The researchers also studied other possible causes for the slow-down but found that repulsion between molecules, variations in molecular shape and "stickiness" between molecules could not account for the dramatic reduction in diffusion rate.
Though the findings are interesting in themselves, their real importance may be in setting the stage for larger studies that would include the thousands of molecules known to be important to cellular operations. Researchers ultimately hope to model everything happening in the cell, including interactions with the cell membrane.
"This is the beginning of what will be a very complicated effort to develop the tools and approaches that will allow us to simulate a sufficiently useful caricature of a cell," Skolnick said. "From that, we will be able to learn the biological principles at work, and then study some 'what if' scenarios."
Those "what if" questions might one day help drug designers better understand how therapeutic compounds work within cells, for instance, or allow cancer researchers to see how cells change from a healthy state to a disease state.
"It would be great if we could study new drugs in a model set of cells to very quickly see what might be the side-effects and cross interactions to understand how we might minimize these problems," Skolnick noted. "The nice thing about a computer simulation is that if it is a reasonably faithful caricature, you can ask a lot of questions – and get answers that help you understand what's going on."
Source: http://gtresearchnews.gatech.edu/