Jun 22 2017
Biomedical Researchers have increased the resolution of their microscopes in order to obtain an in depth understanding of the functions of molecules within the body’s cells. However, these Researchers lack the ability to simultaneously visualize the wide variety of molecules that mediate complicated molecular processes in a single snap-shot.
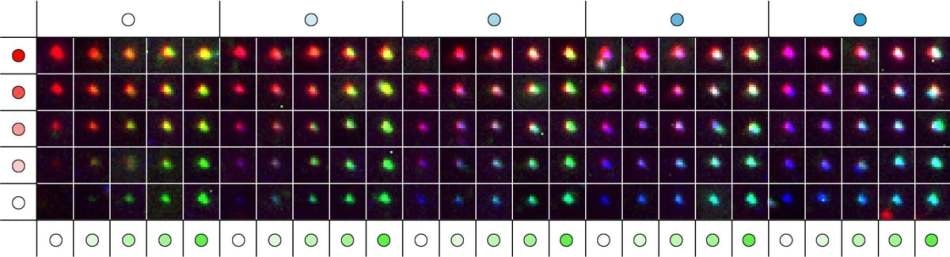 These fluorescence images show a matrix representing 124 distinct metafluorophores, that are generated by combining three fluorescent dyes with varying intensity levels. In the future, the metafluorophore’s unique and identifiable color patterns can be used to analyze the molecular components of complex samples. Credit: Wyss Institute at Harvard University
These fluorescence images show a matrix representing 124 distinct metafluorophores, that are generated by combining three fluorescent dyes with varying intensity levels. In the future, the metafluorophore’s unique and identifiable color patterns can be used to analyze the molecular components of complex samples. Credit: Wyss Institute at Harvard University
A team from Harvard’s Wyss Institute for Biologically Inspired Engineering, the LMU Munich and the Max Planck Institute of Biochemistry in Germany, has recently engineered extremely versatile metafluorophores by incorporating commonly used small fluorescent probes into self-folding DNA structures where their brightness and colors can be digitally programmed. This nanotechnological approach provides a palette of 124 virtual colors for microscopic imaging or various other analytical methods that can be used in the future for visualizing multiple molecular players simultaneously with ultra-high definition. Science Advances has published a report on this method.
The Researchers use this new method to address the problem that until now only a specific number of molecular species can be visualized in a simultaneous manner with fluorescence microscopy in a clinical or biological sample. This problem was solved by introducing fluorescent DNA nanostructures known as metafluorophores, versatile fluorescent dyes whose colors are established based on the manner in which their individual components are arranged in three-dimensional structures.
We use DNA nanostructures as molecular pegboards: by functionalizing specific component strands at defined positions of the DNA nanostructure with one of three different fluorescent dyes, we achieve a broad spectrum of up to 124 fluorescent signals with unique color compositions and intensities. Our study provides a framework that allows researchers to construct a large collection of metafluorophores with digitally programmable optical properties that they can use to visualize multiple targets in the samples they are interested in.
Yin, Core Faculty Member, Wyss Institute and Professor of Systems Biology, Harvard Medical School
It is possible to use the DNA nanostructure-based approach like a barcoding system in order to visually profile the presence of many specific RNA or DNA sequences in samples in what is called multiplexing.
The Researchers took additional engineering steps in order to enable the visualization of multiple molecular structures in tissue samples whose thickness can restrict the movement of bigger DNA nanostructures and make it complicated for them to detect their targets, and to reduce the possibility that they fix themselves to non-specific targets developing false fluorescence signals.
We developed a triggered version of our metafluorophore that dynamically self-assembles from small component strands that take on their prescribed shape only when they bind their target. These in-situ assembled metafluorophores can not only be introduced into complex samples with similar combinatorial possibilities as the prefabricated ones to visualize DNA, but they could also be leveraged to label antibodies as widely used detection reagents for proteins and other biomolecules.
Ralf Jungmann, Ph.D., Faculty at the LMU Munich and the Max Planck Institute of Biochemistry
“This new type of programmable, microscopy-enhancing DNA nanotechnology reveals how work in the Wyss Institute’s Molecular Robotics Initiative can invent new ways to solve long-standing problems in biology and medicine. These metafluorophores that can be programmed to self-assemble when they bind their target, and that have defined fluorescent barcode readouts, represent a new form of nanoscale devices that could help to reveal complex, multi-component, biological interactions that we know exist but have no way of studying today,” said Wyss Founding Director Donald Ingber, M.D., Ph.D., who is also the Judah Folkman Professor of Vascular Biology at Harvard Medical School and the Vascular Biology Program at Boston Children’s Hospital, and Professor of Bioengineering at the Harvard John A. Paulson School of Engineering and Applied Sciences.
Besides Yin and Jungmann, additional Authors on the publication include Johannes Woehrstein and Maximilian Strauss, both members of Jungmann’s research group, and Luvena Ong, Ph.D., Bryan Wei, Ph.D. and David Zhang, Ph.D. who are former members of the Yin group at the Wyss Institute.