Aug 23 2018
In a revolutionary study, an LMU research team headed by Ralf Jungmann has shown that the application of chemically-modified DNA aptamers as protein markers enables one to improve the power of super-resolution fluorescence microscopy as an imaging instrument.
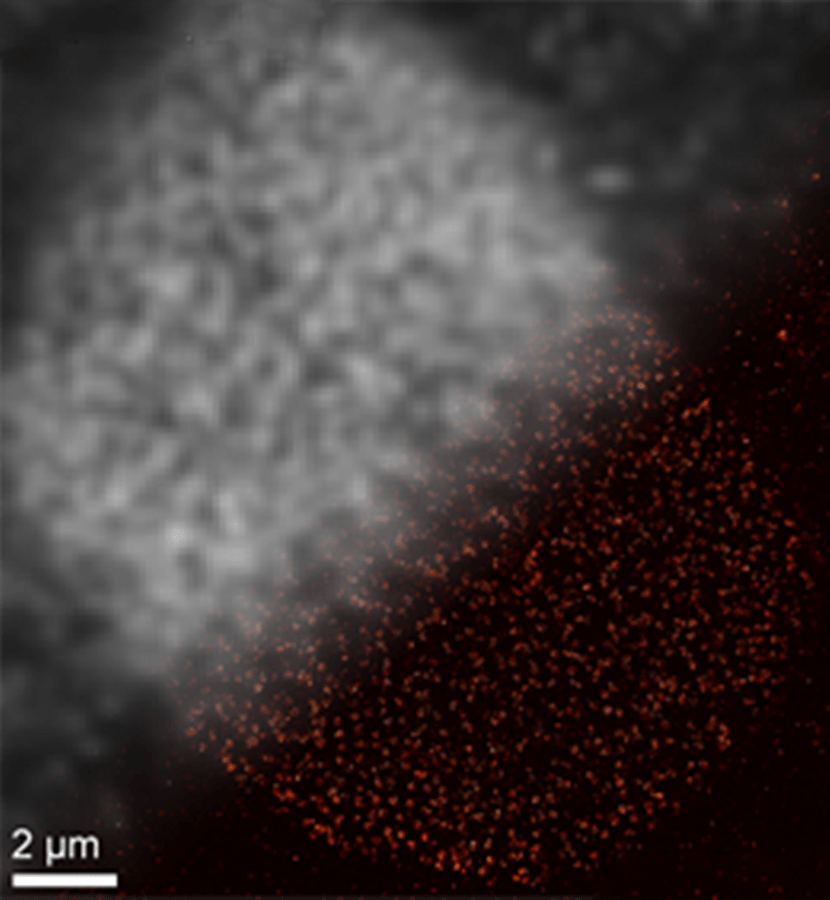 SOMAmer-based DNA-PAINT super-resolution microscopy enables improved spatial resolution. Upper left: Traditional diffraction-limited image of nulcear pore complex proteins on a nuclear cell membrane. Lower right: DNA-PAINT super-resolution image obtained with SOMAmer binders. (Jungmann/LMU)
SOMAmer-based DNA-PAINT super-resolution microscopy enables improved spatial resolution. Upper left: Traditional diffraction-limited image of nulcear pore complex proteins on a nuclear cell membrane. Lower right: DNA-PAINT super-resolution image obtained with SOMAmer binders. (Jungmann/LMU)
Ralf Jungmann has a keen interest in processes that occur within unbelievably tiny spatial dimensions. Holding a professorship in Experimental Physics at LMU, Jungmann heads a research team in Molecular Imaging and Bionanotechnology at the Max Planck Institute for Biochemistry (Martinsried) and concentrates on widening the capabilities of optical microscopy. His aim is to directly view the molecular interactions that occur within individual cells.
In an effort to track the protein networks that occur in such processes, Jungmann employs short DNA strands that are covalently linked to numerous fluorescence markers as probes to find proteins of interest bearing complementary DNA tags. By manipulating the versatility and sequence-specificity of DNA hybridization in this manner, the distributions of huge numbers of molecules in single cells can be imaged at super-resolution. The integration of DNA sequences with varying fluorescent compounds shows why the method is called DNA-PAINT.
The markers used for detecting targets of biological interest are simply too big, which is a major limitation on the ability of super-resolution fluorescence microscopy. “We are working with an instrumental resolution of less than 10 nanometers. But the fluorescent labels conventionally used to tag proteins are much bigger than that. And this factor has hampered the progress of this whole field of research,” explains Jungmann.
This is what pushed the work described in the latest study, which was published in the journal Nature Methods. In this paper, Jungmann and his coworkers investigate the use of the so-called SOMAmers—a unique class of DNA aptamers—as a means to reduce the size of the markers employed in DNA-PAINT. The word ‘aptamer’ was initially coined to describe single-stranded RNA molecules that easily fold into defined three-dimensional (3D) shapes. These molecules have the ability to detect unique species of proteins. The aptamers developed by Jungmann are single-stranded DNA molecules, which fold into defined 3D shapes that attach to specifically targeted proteins.
The ideal label used to tag proteins efficiently and specifically must fulfil several criteria. It should be as small as possible, and it should bind to targets stoichiometrically to enable precise quantitation. In addition, it would be ideal to synthesize whole libraries of these compounds and rapidly identify suitable markers for the proteins of interest.
Sebastian Strauss, a member of Jungmann‘s group and first author of the new study
In order to evaluate the ability of DNA aptamers, the LMU researchers teamed up with SomaLogic, an American company which had already developed a large range of modified aptamers or SOMAmers for other purposes. These SOMAmers can specifically bind a countless number of different proteins. In the latest study, the Munich team altered a selection of these aptamers for DNA-PAINT and came up with efficient labeling protocols for life and fixed cells. The present study reveals that it is certainly possible to further enhance the resolution achievable with traditional fluorescence labels using these innovative labeling reagents together with DNA-PAINT super-resolution microscopy.
“We expect that the new method will provide a significant boost for super-resolution microscopy, particularly with respect to its range of application in biology,” states Ralf Jungmann. He aims to use DNA-PAINT to concurrently view and track as many proteins and their molecular interactions as possible. In upcoming experiments, he and his team intend to use the novel labeling method for imaging the entire protein networks at high resolution. “We will be able to address biological and biomedical questions that have thus far been experimentally inaccessible.”