Dec 7 2020
Researchers from the University of New South Wales (UNSW) have solved a significant design challenge on the way to regulating the dimensions of so-called DNA nanobots—structures that get assembled on their own from DNA components.
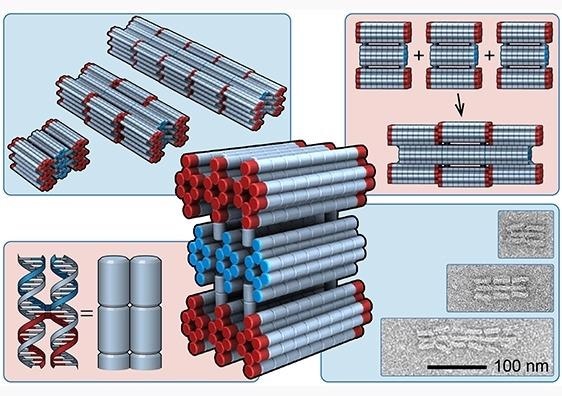 At UNSW, researchers use DNA to build nanorobots. Pictured here are their PolyBricks. Image Credit: Jonathan Berengut.
At UNSW, researchers use DNA to build nanorobots. Pictured here are their PolyBricks. Image Credit: Jonathan Berengut.
Self-assembling nanorobots could seem like science fiction, but a new study in DNA nanotechnology has brought them one step closer to reality. Cases of future use of nanobots will not just be on the small scale but comprise wide-ranging applications in the health and medical field, like wound healing and unblocking of arteries.
Scientists from UNSW, along with collaborators in the United Kingdom, have described a new design theory in the ACS Nano journal on how to regulate the length of self-assembling nanobots without a mold or template.
Traditionally we build structures by manually assembling components into the desired end product. That works quite well and easily if the parts are large, but as you go smaller and smaller, it becomes harder to do this.
Dr Lawrence Lee, Study Lead Author, Single Molecule Science Initiative, UNSW Medicine
Already, medical researchers are capable of building nano-scale robots that could be programmed to perform minuscule tasks, such as to position small electrical components or delivery of drugs to cancer cells.
Scientists at UNSW make use of biological molecules—such as DNA—to develop such nanorobots. Through a process known as molecular self-assembly, small individual component parts build themselves into huge structures.
The difficulty with the use of self-assembly to construct is identifying how to program the building blocks to develop the preferred structure and making them to stop when the structure is tall or long enough.
As part of this project, the UNSW scientists executed their design by producing DNA subunits, known as PolyBricks. As it occurs in natural systems, each of the building blocks is encoded with the master plans to self-assemble into pre-configured structures of set length.
Dr Lee compares the PolyBricks to the microbots in the scifi film Big Hero Six, where the microbots self-assemble into multiple different formations.
In the film, the ultimate robot is a bunch of identical subunits that can be instructed to self-assemble into any desired global form.
Dr Lawrence Lee, Study Lead Author, Single Molecule Science, UNSW Medicine
The researchers employed a design principle called strain accumulation to regulate the dimensions of their built structures.
With each block we add, strain energy accumulates between the PolyBricks, until ultimately the energy is too great for any more blocks to bind. This is the point at which the subunits will stop assembling.
Dr Lawrence Lee, Study Lead Author, Single Molecule Science, UNSW Medicine
To regulate the length of the final structure—that is, how many PolyBricks are combined together—the research team altered the sequence in their DNA design to control how much strain has been added with every new block.
According to Dr Lee, “Our theory could help researchers design other ways to use strain accumulation to control the global dimensions of open self-assemblies.”
The authors state this mechanism could be employed to encode more complex shapes with the help of self-assembly units.
“It’s this type of fundamental research into how we organise matter at the nanoscale that’s going to lead us to the next generation of nanomaterials, nanomedicines, and nanoelectronics,” stated PhD graduate and study lead author, Dr Jonathan Berengut.
Journal Reference
Berengut, J. F., et al. (2020) Self-Limiting Polymerization of DNA Origami Subunits with Strain Accumulation. ACS Nano. doi.org/10.1021/acsnano.0c07696.