Jan 11 2021
In cells, ribosomes are considered as the complexes of ribonucleoproteins central to the synthesis of proteins. But the lack of concrete evidence has made it difficult to understand how such complexes function.
 es function.
es function.
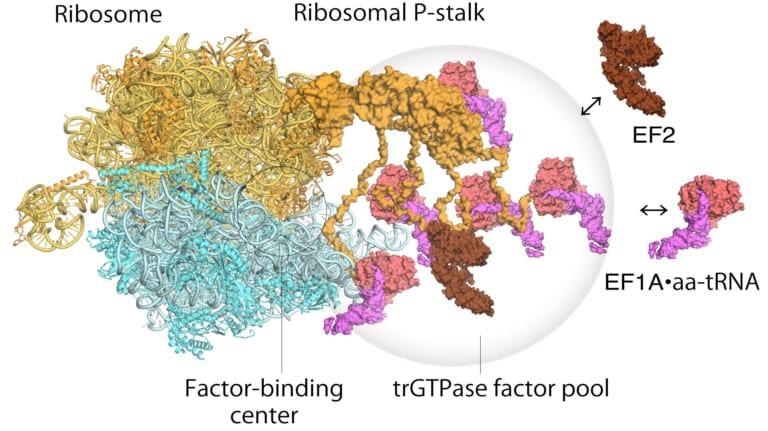
 Model of translating ribosomes and elongation factors. EF1A•GTP•aatRNA and EF2 assemble to the ribosomal stalk on the translating ribosome. The translation factor pool contributes to efficient protein synthesis in a crowded intracellular environment. Image Credit: Proceedings of the National Academy of Sciences.
Model of translating ribosomes and elongation factors. EF1A•GTP•aatRNA and EF2 assemble to the ribosomal stalk on the translating ribosome. The translation factor pool contributes to efficient protein synthesis in a crowded intracellular environment. Image Credit: Proceedings of the National Academy of Sciences.
Hirotatsu Imai and Noriyuki Kodera from Kanazawa University, together with Toshio Uchiumi from Niigata University in Japan, have now achieved visualizations of the structural dynamics and factor pooling that occur at ribosome stalk proteins as they create new proteins.
Discovered first in the 1950s, the wide function of ribosomes has been extensively studied for some time—they read messenger RNA sequences to produce sequences of properly ordered amino acids into new proteins.
In particular, the ribosome stalk protein plays a crucial role in the process of protein synthesis by engaging protein factors that are accountable for elongation and translation of the amino acid sequence. But it has been difficult to set up the structure of the bound ribosome stalk protein in an acceptable way due to its resilience. In this context, the fast image capture and high resolution of the high-speed atomic force microscopy were found to be crucial.
Atomic force microscopy involves using a nanoscale tip to feel samples, quite similar to a vinyl record player needle that scans over a record, only that the resolution of the details recognized by an atomic force microscope can be on an atomic scale.
The flexibility of the method for various surfaces was already highly beneficial for biological studies, but with the emergence of high-speed atomic force microscopy, the technique could capture dynamic processes initially as well. Imai, Uchiumi, and Kodera made use of this method to disclose that the stalk protein flips between two conformations—one that matches with earlier structural models and one completely new conformation that was never anticipated.
As far as the functioning of a ribosome is concerned, a two-step mechanism had been proposed earlier to explain how genetic information is translated via proteins called “translational GTPase factors.” The initial step is the recruitment of the factors to the factor-tethering site on the protein stalk, thus increasing the focus of factors there—known as factor pooling.
The second step is the stabilizing and binding of a translational GTPase on the ribosomal factor, which acts as a binding center to catalyze GTPase hydrolysis. Starting from their high-speed atomic force microscopy study, the scientists were able to get the initial visual evidence for the translational GTPase factor pooling mechanism with the help of ribosomal stalk.
The study failed to provide decisive evidence of the action of the factors once bound, but the team did observe that the factors seemed to be retained in the proximity once GTPase hydrolysis was done. It indicates a possible role of the stalk protein in additional stages of protein synthesis.
The team came up with the conclusion that “future work with HS-AFM will provide further important information to understand the dynamic behaviors of these complex translational machineries.”
Journal Reference
Imai, H., et al. (2020) Direct visualization of translational GTPase factor pool formed around the archaeal ribosomal P-stalk by high-speed AFM. Proceedings of the National Academy of Sciences. doi.org/10.1073/pnas.2018975117.