Bottom-up engineering of synthetic condensate offers an understanding of condensate organization and helps to develop synthetic systems with unprecedented functions. Nevertheless, realizing such systems with predictable organization and desirable functions remains challenging.

Study: Engineering DNA-based synthetic condensates with programmable material properties, compositions, and functionalities. Image Credit: PopTika/Shutterstock.com
A study published in Science Advances presented DNA-based synthetic condensates assembled via phase separation. Modulating the DNA molecule’s intermolecular interactions regulated various properties of condensate, such as composition, assembly, and function. Like intracellular condensates, DNA clients are partitioned preferentially into cognate condensates.
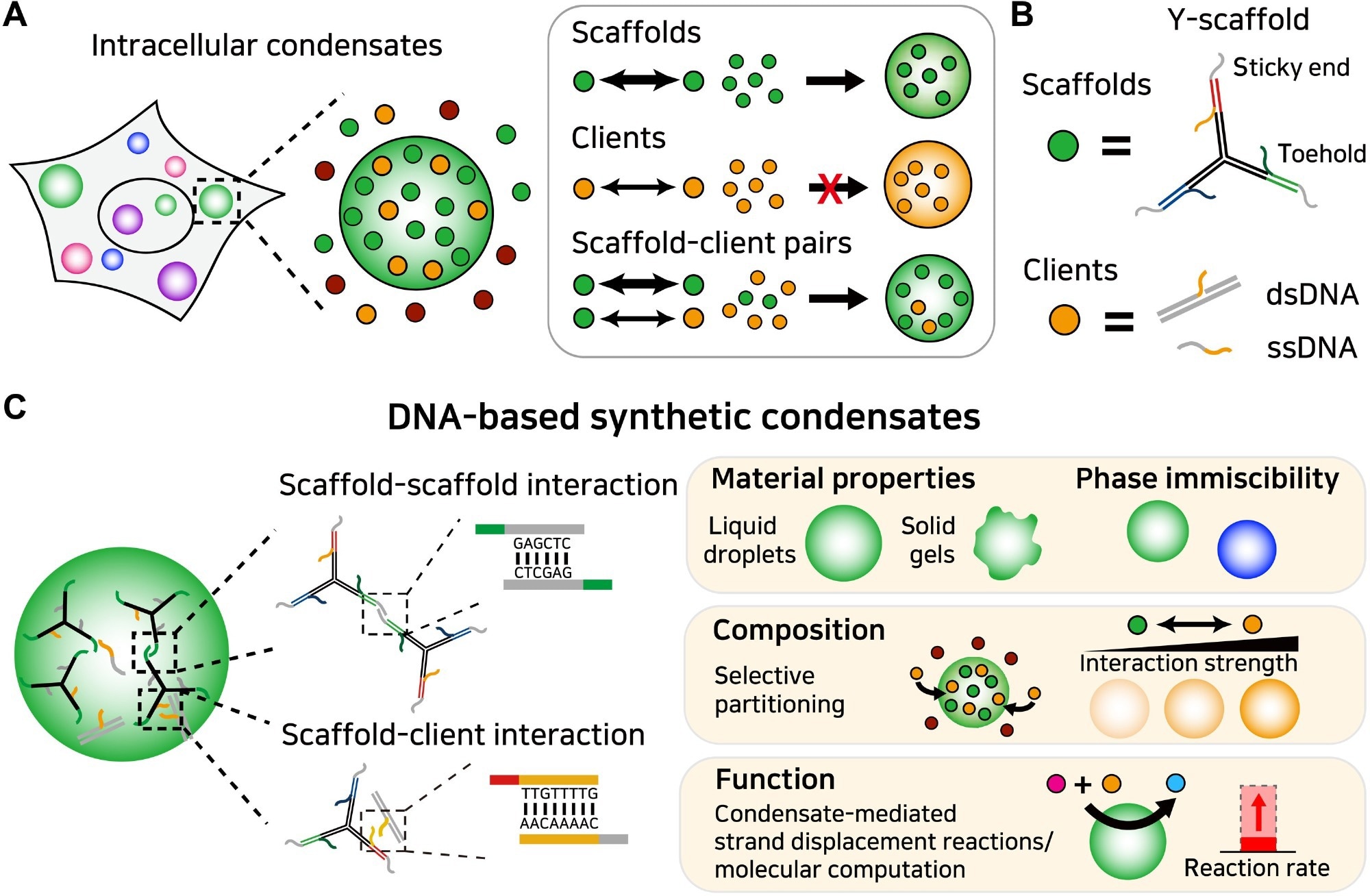
Figure 1. Programmable organization and function of DNA-based synthetic condensates. (A) Intracellular condensates perform cellular functions by concentrating specific biomolecules that can be classified into scaffolds and clients. Scaffolds drive condensate assembly, and clients partition into the condensate. (B) In our DNA-based synthetic condensates, a Y-shaped DNA nanostructure acts as a scaffold. It harbors sticky ends for phase separation and toeholds for composition control. Clients interact with the scaffold via an ssDNA region complementary to the toehold. (C) The organization and function of DNA-based synthetic condensates. Using the sequence programmability of DNA, the strengths of intermolecular interactions among condensate components can be precisely defined. This feature enables control over multiple condensate properties including phase behaviors, compositions, and functions. Image Credit: Do, S et al.,Science Advances
Concentrating specific reaction components enabled synthetic condensates to expedite logic gate operations and DNA strand displacement reactions. Thus, DNA-based condensates helped achieve the high-order functionalities required to create life-like artificial systems.
Intracellular Condensates
Intracellular condensates include classic membrane-less organelles, such as nuclear speckles and nucleoli, and recently identified structures, including synaptic, transcriptional, and signaling clusters. Each condensate performs various cellular activities, primarily by modulating the composition of its constituents.
Biomolecule phase separation has surfaced as a basic principle of intracellular organization. Despite minimal research, a network of transitory multivalent intermolecular interactions has been identified to determine the content and organization of condensates.
The complexity of protein-protein interactions frequently impedes a thorough understanding of intermolecular interactions at the nanoscale level, generating multiphase behaviors in micrometer-scale condensates.
As a building block, DNA has distinct advantages over all other known materials because of its specific intermolecular interactions and thermodynamic stability, which can be precisely determined from its linear sequence.
The outstanding programmability of DNA molecules has been widely established in various situations where DNA strand combinations self-assemble into sophisticated three-dimensional (3D) structures. DNA sequence programming was originally used to produce static assemblies but has also been used to generate dynamic assemblies.
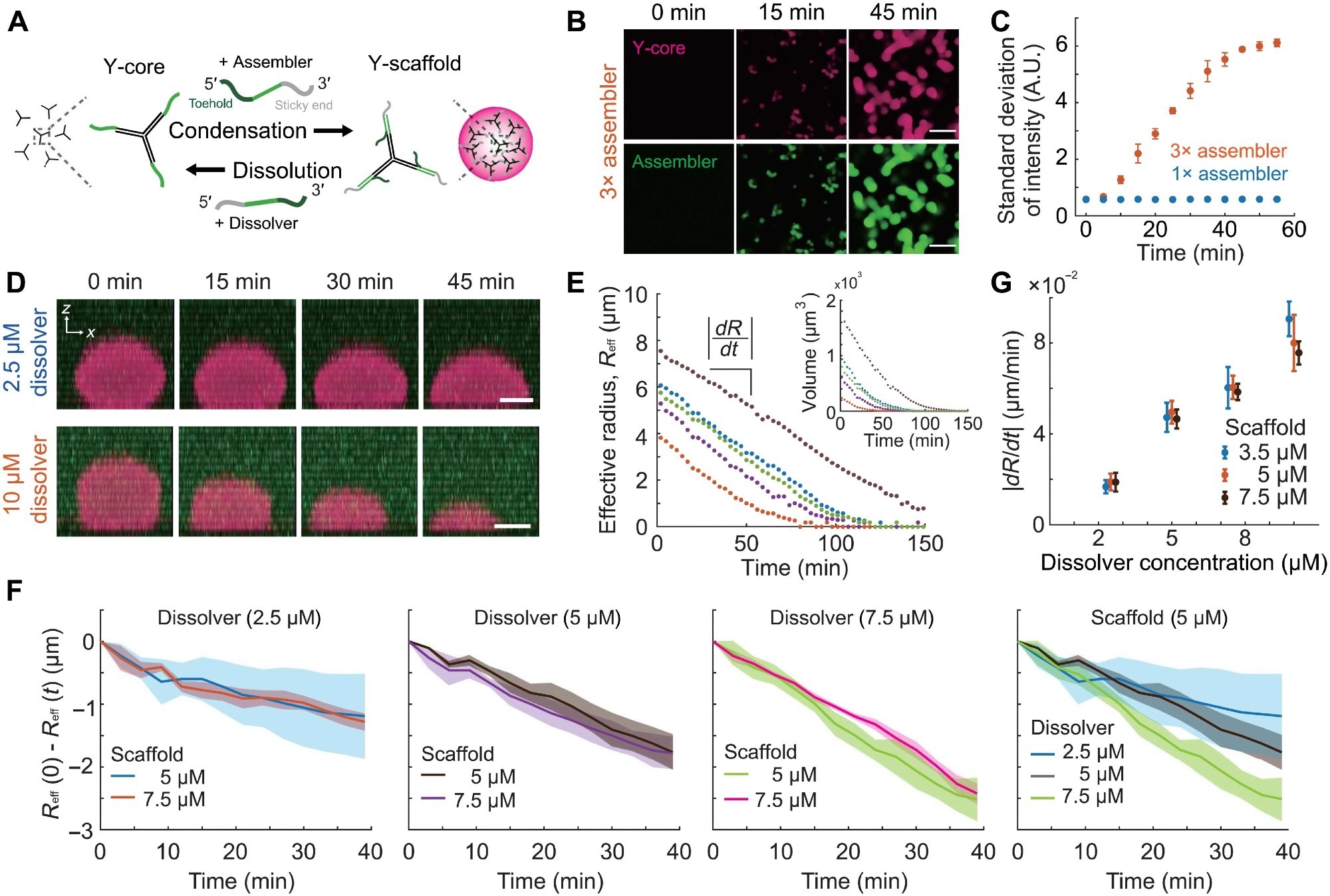
Figure 2. The control of condensation and dissolution of the DNA condensate. (A) Schematic of dynamic condensation and dissolution of the Y-scaffold. Binding of the assembler to the Y-core leads to the formation of the trivalent Y-scaffold, triggering phase separation. Upon addition of the dissolver, the toehold-mediated strand displacement reaction strips off the assembler from the Y-core, resulting in condensate dissolution. (B) Time-lapse images of dynamic condensation experiments with different concentrations of the assembler. Y-core (10 μM) is present in the chamber before the addition of 30 μM assembler. (C) Temporal changes of the SDs of the assembler channel images in (B). Identical quantification on the 10 μM assembler data is shown as well. (D) Time-lapse images of the side view of condensate dissolution obtained with different dissolver concentrations. Before the addition of dissolvers, condensates assembled at 5 μM Y-scaffolds are present in the sample. (E) Temporal changes of the effective radius of condensates, Reff, and condensate volumes (inset). Dissolvers (5 μM) are added to the sample with equimolar Y-scaffolds. (F) The temporal changes of the effective radius of condensates measured with different concentrations of scaffolds and dissolvers. For each condition, the average and the SD of five different condensates are plotted. All data were acquired at 30°C and 350 mM NaCl. (G) Condensate dissolution rates obtained with different concentrations of scaffolds and dissolvers. Dissolver concentrations used are 2.5, 5, 7.5, and 10 μM. Data points are shifted slightly in the x axis for clarity. Scale bars, 20 μm (B) and 5 μm (D). Image Credit: Do, S et al.,Science Advances
The discovery of displacement reactions in DNA strands has opened new possibilities for a broad range of applications requiring a rapid exchange of individual DNA strands, such as molecular computing, programmable hydrogels, and automated nanomachines.
Watson-Crick base pairing-based biomolecular phase separation has been linked to the formation of intracellular condensates and has also been demonstrated with DNA nanostructures of restricted valency, known as DNA nanostars.
Each arm’s sticky end mediates self-association in DNA nanostars, a property that is extremely valuable for studying macroscale rheological features and thermodynamics as well as for analyzing the relationship between intermolecular interactions. The phase behaviors of DNA nanostar systems are characterized by low valence-associated contraction of the coexistence zone and the formation of an equilibrium gel.
DNA-Based Synthetic Condensates
The phase separation mechanism in cells helps to organize their contents by forming membrane-less condensates. Specific biomolecules incorporated into condensates interact with each other to perform various biological processes.
This study presented DNA-based synthetic condensates that mimic the organization and function of natural intracellular condensates. Inspired by the unique properties of DNA in programming intermolecular interactions, DNA strands of clients and scaffolds have been constructed with the ability to undergo self-assembly to form functional condensates. The self-association scaffolds were built from well-characterized Y-shaped DNA nanostars.
The current study showed that preferential partitioning and enrichment of clients in condensates significantly expedited displacement reactions in DNA strands, a property used in molecular computation. Thus, the DNA condensates in this study offered a programmable platform in which their function and composition can be predicted.
The primary goal of this study was to demonstrate the rational design of DNA-based synthetic condensates. Although a three-arm DNA scaffold was employed because of its facile implementation and high sensitivity to manipulation, higher-valence scaffolds can produce condensates with distinct physical characteristics.
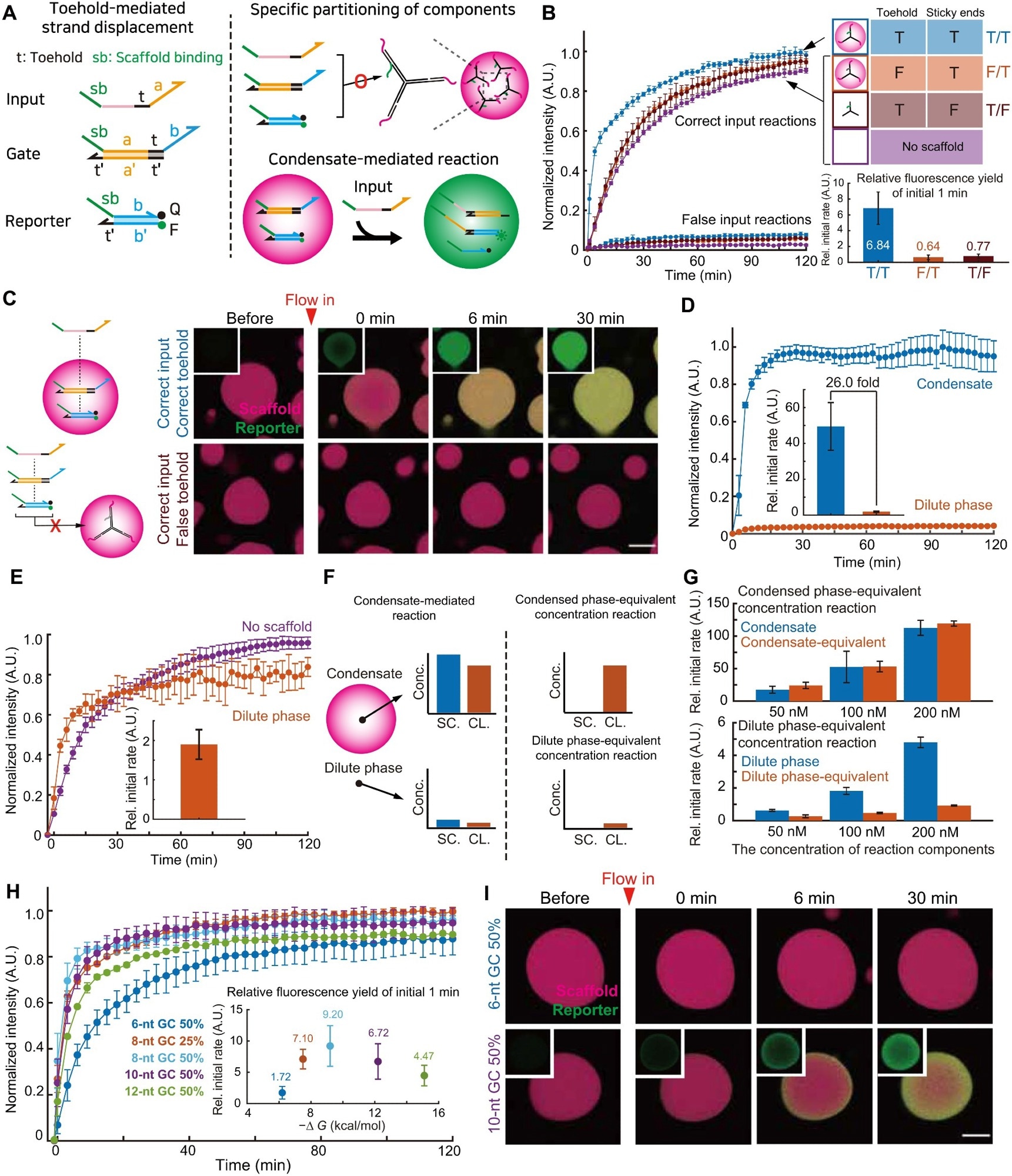
Figure 3. Quantitative analysis of condensate mediated strand displacement reactions. (A) Schematics of a double-cascade DNA strand displacement reaction used in this figure. (B) Left: Kinetics of the reactions shown in (A) with either different inputs or scaffolds, measured with a plate reader. “T” and “F” indicate true and false sequences, respectively. (C) Fluorescence images of the condensate-mediated strand displacement reactions. (D) Kinetics of the reactions in the dilute and the dense phase, obtained from imaging data as in (C). Error bars are SDs (three ROIs per sample; three independently prepared samples) (E) Kinetics of strand displacements in the dilute phase of the two-phase reaction and the single-phase one in the absence of scaffolds. (F) Schematic of the experiments to probe the contribution of the mass action in the condensate-mediated reaction. “SC.” and “CL.” indicate scaffolds and clients, respectively. (G) The results of experiments designed in (F) at indicated client concentrations. All data are normalized with the initial reaction rate in the absence of scaffolds at the client concentrations of 100 nM. Error bars are SDs (n = 3 independent experiments). (H) The reaction kinetics conducted with client-scaffold pairs of different binding strengths. The inset shows the normalized reaction rates during the initial 1 min plotted against the hybridization free energy between scaffolds and clients. Error bars are SDs (n = 3 independent experiments). (I) Time-lapse images of the condensate-mediated reactions conducted with scaffold-client pairs of different binding strengths. Scale bars, 20 μm (C and I). In this figure, unless otherwise specified, all reaction components are at 100 nM and have a scaffold binding site of 8-nt GC 25%. All the relative initial rates in the insets of this figure show the fluorescence increase over initial 1 min normalized with the corresponding value in the absence of scaffolds. Image Credit: Do, S et al.,Science Advances
The binodals for the condensate assembly increased with valency, resulting in a shift in the viscoelastic characteristics, and enabling condensates to operate at higher temperatures or across a wide concentration range with possibly differing reaction rates. Furthermore, the method described herein could be used to create multimodal condensates.
Like natural condensates, synthetic DNA condensates, may emit signals that can, for instance, act on other DNA circuits in solution or condensates. It is also possible to create synthetic condensates that function in the sequestration process by depleting a pool of certain components.
Conclusion
In summary, a DNA-based synthetic condensate platform was designed with the ability to tune the composition and function of condensates from individual sequences. This study demonstrated that the dissolution and condensation of condensates could be regulated by specific signals acting on the scaffolds.
In the future, researchers hope that DNA-based condensate platforms will provide a layer of high-order functionalities that are not possible with diluted biomolecules. It is anticipated that the unique properties of DNA (intermolecular interaction sequence programmability, biocompatibility, and others) will make it a crucial component for the development of increasingly life-like synthetic systems.
Reference
Do, S., Lee, C., Lee, T., Kim, D. N., Shin, Y. (2022). Engineering DNA-based synthetic condensates with programmable material properties, compositions, and functionalities. Science Advances. https://www.science.org/doi/10.1126/sciadv.abj1771
Disclaimer: The views expressed here are those of the author expressed in their private capacity and do not necessarily represent the views of AZoM.com Limited T/A AZoNetwork the owner and operator of this website. This disclaimer forms part of the Terms and conditions of use of this website.