Sep 28 2015
A team led by researchers at UC Santa Cruz has developed chip-based technology for reliable detection of Ebola virus and other viral pathogens. The system uses direct optical detection of viral molecules and can be integrated into a simple, portable instrument for use in field situations where rapid, accurate detection of Ebola infections is needed to control outbreaks.
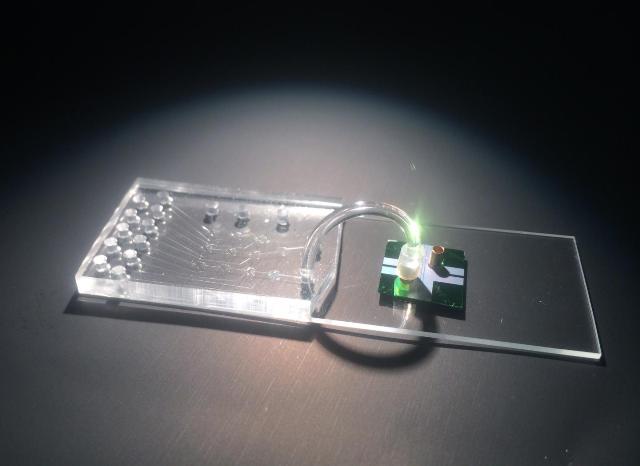 This hybrid device integrates a microfluidic chip for sample preparation and an optofluidic chip for optical detection of individual molecules of viral RNA. Credit: Joshua Parks
This hybrid device integrates a microfluidic chip for sample preparation and an optofluidic chip for optical detection of individual molecules of viral RNA. Credit: Joshua Parks
Laboratory tests using preparations of Ebola virus and other hemorrhagic fever viruses showed that the system has the sensitivity and specificity needed to provide a viable clinical assay. The team reported their results in a paper published September 25 in Nature Scientific Reports.
An outbreak of Ebola virus in West Africa has killed more than 11,000 people since 2014, with new cases occurring recently in Guinea and Sierra Leone. The current gold standard for Ebola virus detection relies on a method called polymerase chain reaction (PCR) to amplify the virus's genetic material for detection. Because PCR works on DNA molecules and Ebola is an RNA virus, the reverse transcriptase enzyme is used to make DNA copies of the viral RNA prior to PCR amplification and detection.
"Compared to our system, PCR detection is more complex and requires a laboratory setting," said senior author Holger Schmidt, the Kapany Professor of Optoelectronics at UC Santa Cruz. "We're detecting the nucleic acids directly, and we achieve a comparable limit of detection to PCR and excellent specificity."
In laboratory tests, the system provided sensitive detection of Ebola virus while giving no positive counts in tests with two related viruses, Sudan virus and Marburg virus. Testing with different concentrations of Ebola virus demonstrated accurate quantification of the virus over six orders of magnitude. Adding a "preconcentration" step during sample processing on the microfluidic chip extended the limit of detection well beyond that achieved by other chip-based approaches, covering a range comparable to PCR analysis.
"The measurements were taken at clinical concentrations covering the entire range of what would be seen in an infected person," Schmidt said.
Schmidt's lab at UC Santa Cruz worked with researchers at Brigham Young University and UC Berkeley to develop the system. Virologists at Texas Biomedical Research Institute in San Antonio prepared the viral samples for testing.
The system combines two small chips, a microfluidic chip for sample preparation and an optofluidic chip for optical detection. For over a decade, Schmidt and his collaborators have been developing optofluidic chip technology for optical analysis of single molecules as they pass through a tiny fluid-filled channel on the chip. The microfluidic chip for sample processing can be integrated as a second layer next to or on top of the optofluidic chip.
Schmidt's lab designed and built the microfluidic chip in collaboration with coauthor Richard Mathies at UC Berkeley who pioneered this technology. It is made of a silicon-based polymer, polydimethylsiloxane (PDMS), and has microvalves and fluidic channels to transport the sample between nodes for various sample preparation steps. The targeted molecules--in this case, Ebola virus RNA--are isolated by binding to a matching sequence of synthetic DNA (called an oligonucleotide) attached to magnetic microbeads. The microbeads are collected with a magnet, nontarget biomolecules are washed off, and the bound targets are then released by heating, labeled with fluorescent markers, and transferred to the optofluidic chip for optical detection.
Schmidt noted that the team has not yet been able to test the system starting with raw blood samples. That will require additional sample preparation steps, and it will also have to be done in a biosafety level 4 facility.
"We are now building a prototype to bring to the Texas facility so that we can start with a blood sample and do a complete front-to-back analysis," Schmidt said. "We are also working to use the same system for detecting less dangerous pathogens and do the complete analysis here at UC Santa Cruz."