Aug 16 2016
 According to researchers at Rice University, graphene nanoribbons (GNRs) can be adapted for biological applications, such as drug delivery, DNA analysis, and biomimetic applications, as they bend and twist easily in solution.
According to researchers at Rice University, graphene nanoribbons (GNRs) can be adapted for biological applications, such as drug delivery, DNA analysis, and biomimetic applications, as they bend and twist easily in solution.
According to researchers at Rice University, graphene nanoribbons (GNRs) can be adapted for biological applications, such as drug delivery, DNA analysis, and biomimetic applications, as they bend and twist easily in solution.
Understanding the behavior of GNRs in a solution will help to make this material suitable for extensive use in biomimetics, explains Rice physicist Ching-Hwa Kiang, whose lab used its unique features to investigate nanoscale materials such as proteins and cells in wet environments. Biomimetic materials are those that mimic the properties and forms of natural materials.
The research was headed by recent Rice graduate Sithara Wijeratne, now a postdoctoral researcher at Harvard University, and the research paper was published in the Nature journal Scientific Reports.
GNRs tend to be several times longer than they are wide. They can be generated in bulk by chemically “unzipping” carbon nanotubes, a method formulated by Rice chemist and co-author James Tour and his lab.
Their size indicates that they can function on the scale of biological components, such as DNA and proteins, Kiang explained.
We study the mechanical properties of all different kinds of materials, from proteins to cells, but a little different from the way other people do. We like to see how materials behave in solution, because that’s where biological things are.
Ching-Hwa Kiang, Physicist, Rice University
Kiang is a pioneer in inventing techniques to explore the energy states of proteins as they fold and unfold.
She said it was Tour’s idea for her lab to look at the mechanical properties of GNRs. “It’s a little extra work to study these things in solution rather than dry, but that’s our specialty,” she said.
It is a known fact that nanoribbons can add strength but not weight to solid-state composites - such as tennis rackets and bicycle frames - and produce an electrically active matrix.
An earlier Rice project has infused them into an efficient de-icer coating designed for aircraft. But in a squishier environment, their ability match surfaces, transmit current and strengthen composites could also be crucial.
It turns out that graphene behaves reasonably well, somewhat similar to other biological materials. But the interesting part is that it behaves differently in a solution than it does in air.
Ching-Hwa Kiang, Physicist, Rice University
The team discovered that similar to proteins and DNA, nanoribbons in a solution naturally form loops and folds, but can also develop wrinkles, helicoids, and spirals.
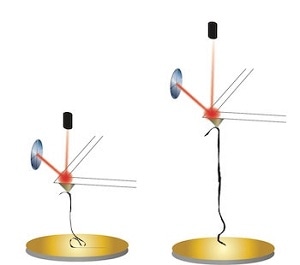
Kiang, Wijeratne and Jingqiang Li, a co-author and student in the Kiang lab, used atomic force microscopy to examine their properties. Atomic force microscopy is capable of collecting high-resolution images as well as obtaining sensitive force measurements of nanomaterials by pulling on them. The researchers investigated GNRs and their precursors, graphene oxide nanoribbons.
The Rice team noticed that all nanoribbons became stiff under stress, but their stiffness grows as oxide molecules are eliminated to transform graphene oxide nanoribbons into GNRs. They recommended this ability to tweak their stiffness, which would aid in the design and fabrication of GNR-biomimetic interfaces.
Graphene and graphene oxide materials can be functionalized (or modified) to integrate with various biological systems, such as DNA, protein and even cells. These have been realized in biological devices, biomolecule detection and molecular medicine. The sensitivity of graphene bio-devices can be improved by using narrow graphene materials like nanoribbons.
Ching-Hwa Kiang, Physicist, Rice University
Wijeratne observed that GNRs were being examined for use in DNA sequencing, where strands of DNA are pulled via a nanopore present in an electrified material. The DNA’s base components impact the electric field, which can be read to detect the bases.
The biocompatibility of nanoribbons was noticed by the researchers to possess the potential use for sensors that could travel through the body and provide information on what they discover, quite similar to the nanoreporters built by Tour lab’s that recover data from oil wells.
Going forward, the research will focus on the effect of the width of the nanoribbons, which measures from 10 to 100 nm, on their properties.
Co-authors are Rice research scientist Evgeni Penev; graduate student Wei Lu; alumna Amanda Duque, now a scientist at Los Alamos National Laboratory; and Boris Yakobson, the Karl F. Hasselmann Professor of Materials Science and NanoEngineering and a professor of chemistry. Tour is the T.T. and W.F. Chao Professor of Chemistry as well as a professor of computer science and of materials science and nanoengineering. Kiang is an associate professor of physics and astronomy and of bioengineering.
The research was supported by the Welch Foundation and the National Science Foundation. The researchers used the NSF’s Extreme Science and Engineering Discovery Environment and the NSF-supported DAVinCI supercomputer administered by Rice’s Center for Research Computing and procured in a partnership with Rice’s Ken Kennedy Institute.