Nov 30 2018
Several different microbes, such as viruses and bacteria, could be responsible for causing pneumonia, a respiratory disease that takes the lives of nearly 50,000 people in the United States each year.
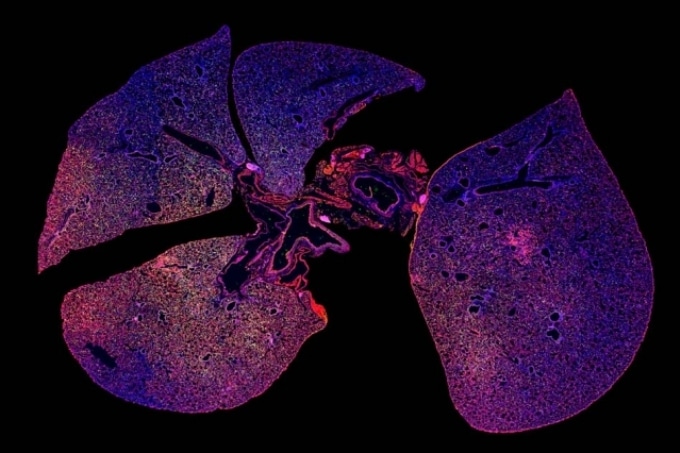 A strong immune response can be seen in this immunofluorescence image of lung tissue infected with pneumonia where immune cells are stained green and red. (Image credit: Colin Buss)
A strong immune response can be seen in this immunofluorescence image of lung tissue infected with pneumonia where immune cells are stained green and red. (Image credit: Colin Buss)
Quick diagnosis of pneumonia is crucial for effective treatment, specifically in hospital-acquired cases which are usually more severe. However, existing diagnostic strategies generally need a number of days to return definitive results, rendering it more difficult for doctors to prescribe the right treatment.
At present, scientists at MIT have created a nanoparticle-based technology that could be applied to enhance the speed of diagnosis. This kind of sensor could even be applied to monitor whether or not antibiotic therapy has effectively treated the infection, stated Sangeeta Bhatia, the John and Dorothy Wilson Professor of Health Sciences and Technology and Electrical Engineering and Computer Science and the senior author of the study, who is also a member of MIT’s Koch Institute for Integrative Cancer Research and Institute for Medical Engineering and Science.
If the patient’s symptoms go away, then you assume the drug is working. But if the patient’s symptoms don’t go away, then you would want to see if the bacteria is still growing. We were trying to address that issue.
Sangeeta Bhatia, Professor, MIT.
The lead authors of the study, which was published online in the EBioMedicine journal on November 29th, 2018, are graduate student Colin Buss and recent PhD recipient Jaideep Dudani. Other authors of the paper are Reid Akana, an MIT senior, and Heather Fleming, director of research for Bhatia’s lab.
Sensors in the body
Many years back, Bhatia and her team devised a diagnostic strategy that involves amplifying a signal from biomarkers already present in the body—especially, enzymes known as proteases, which chop up other proteins. The human genome encodes over 500 different proteases, each of which targets distinct proteins.
The researchers created nanoparticles coated with peptides, or short proteins, which can be chopped up by specific proteases, for instance, those expressed by cancer cells. Upon being injected into the body, these particles get accumulated in tumors, if any exist, and proteases present there chop the peptides from the nanoparticles. These peptides are removed in the form of waste and can be detected through a simple urine test.
We’ve been working on this idea that measuring enzyme activity could be a new way to peer inside the body.
Sangeeta Bhatia, Professor, MIT.
She has demonstrated in recent studies that this strategy can be applied for the detection of different types of cancers, such as very small ovarian tumors, which could allow earlier diagnosis of ovarian cancer.
For its new research, the team intended to examine the possibility of diagnosing infection through the detection of proteases produced by microbes. They started with a species of bacteria known as Pseudomonas aeruginosa, which can cause pneumonia and is specifically a common cause of hospital-acquired cases. Since Pseudomonas expresses a protease known as LasA, the team designed nanoparticles with peptides that can be cleaved by LasA.
The team also created a second nanoparticle-based sensor with the ability to monitor the immune response of the host to infection. These nanoparticles are covered in peptides that are cleaved by a kind of protease known as elastase, which is formed by immune cells known as neutrophils.
In certain patients suffering from pneumonia, even if the bacteria causing the infection is cleared out by an antibiotic, a chest X-ray might still reveal inflammation since neutrophils are still active. When the two sensors are used together, it could be found whether an antibiotic has cleared the infection, in cases where a chest X-ray still reveals inflammation after treatment.
The sensors can help you distinguish between whether there’s an infection and inflammation, versus inflammation and no infection. What we showed in the paper is that when you treat with the right antibiotic, the infection goes down but the inflammation persists.
Sangeeta Bhatia, Professor, MIT.
The team also demonstrated that when mice were treated with an ineffective antibiotic, both inflammation levels and bacteria levels remained high. This type of test could help reveal whether an antibiotic works, in cases where the symptoms of a patient have not improved within a few days.
Diagnosing many infections
As part of this study, the team intravenously delivered the nanoparticles; however, they are now working on a powdered version that is safe to be inhaled.
Bhatia visualizes that this strategy could be applied to identify whether a patient has viral or bacterial pneumonia, which would be assistive to doctors in deciding whether or not the patient should be administered with antibiotics. Since it takes several days for the definitive test, that is, growing a bacterial culture from coughed-up mucus, doctors base their decisions on the symptoms of the patients and X-ray imaging—a process that may not always be precise.
In order to develop a more comprehensive diagnostic, currently, Bhatia’s lab is striving to add peptides with the potential to interact with proteases from other kinds of bacteria that cause pneumonia, and even proteases produced by the host immune system in response to bacterial or viral infection. The researchers are also working on sensors with the capability of easily differentiating between active and dormant forms of tuberculosis.
Bhatia and her colleagues have started a company known as Glympse Bio that has licensed the protease sensing technology and is now striving to develop protease sensors for possible use in humans. Next year, they intend to start a phase I clinical trial of a sensor with the potential to detect liver fibrosis, an accumulation of scar tissue that can cause cirrhosis.
The Koch Institute Support (core) Grant from the National Cancer Institute, the Core Center Grant from the National Institute of Environmental Health Sciences, and the National Institute of Allergy and Infectious Diseases funded the study.