Oct 27 2014
Adherent cells, the kind that form the architecture of all multi-cellular organisms, are mechanically engineered with precise forces that allow them to move around and stick to things. Proteins called integrin receptors act like little hands and feet to pull these cells across a surface or to anchor them in place. When groups of these cells are put into a petri dish with a variety of substrates they can sense the differences in the surfaces and they will "crawl" toward the stiffest one they can find.
 Our premise is that mechanics play a role in almost all biological processes, and with these DNA-based tension probes we're going to uncover, measure and map those forces,' says biomolecular chemist Khalid Salaita. Credit: Graphic by Victor Ma.
Our premise is that mechanics play a role in almost all biological processes, and with these DNA-based tension probes we're going to uncover, measure and map those forces,' says biomolecular chemist Khalid Salaita. Credit: Graphic by Victor Ma.
Now chemists have devised a method using DNA-based tension probes to zoom in at the molecular level and measure and map these phenomena: How cells mechanically sense their environments, migrate and adhere to things.
Nature Communications published the research, led by the lab of Khalid Salaita, assistant professor of biomolecular chemistry at Emory University. Co-authors include mechanical and biological engineers from Georgia Tech.
Using their new method, the researchers showed how the forces applied by fibroblast cells is actually distributed at the individual molecule level. "We found that each of the integrin receptors on the perimeter of cells is basically 'feeling' the mechanics of its environment," Salaita says. "If the surface they feel is softer, they will unbind from it and if it's more rigid, they will bind. They like to plant their stakes in firm ground."
Each cell has thousands of these integrin receptors that span the cellular membrane. Cell biologists have long been focused on the chemical aspects of how integrin receptors sense the environment and interact with it, while the understanding of the mechanical aspects lagged. Cellular mechanics is a relatively new but growing field, which also involves biophysicists, engineers, chemists and other specialists.
"Lots of good and bad things that happen in the body are mediated by these integrin receptors, everything from wound healing to metastatic cancer, so it's important to get a more complete picture of how these mechanisms work," Salaita says.
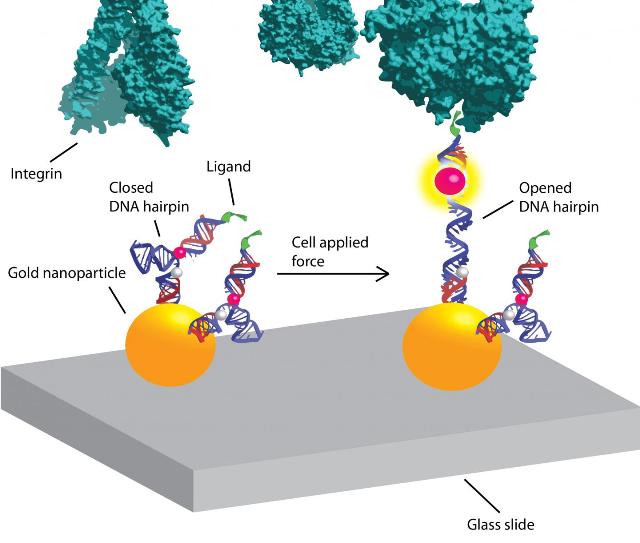
The Salaita lab previously developed a fluorescent-sensor technique to visualize and measure mechanical forces on the surface of a cell using flexible polymers that act like tiny springs. These springs are chemically modified at both ends. One end gets a fluorescence-based turn-on sensor that will bind to an integrin receptor on the cell surface. The other end is chemically anchored to a microscope slide and a molecule that quenches fluorescence. As force is applied to the polymer spring, it extends. The distance from the quencher increases and the fluorescent signal turns on and grows brighter. Measuring the amount of fluorescent light emitted determines the amount of force being exerted.
Yun Zhang, a co-author of the Nature Communications paper and a graduate student in the Salaita lab, had the idea of using DNA molecular beacons instead of flexible polymers. "She was new to the lab and brought a fresh perspective," Salaita says.
The molecular beacons are short pieces of lab-synthesized DNA, each consisting of about 20 base pairs, used in clinical diagnostics and research. The beacons are called DNA hairpins because of their shape.
The thermodynamics of DNA, its double-strand helix structure and the energy needed for it to fold are well understood, making the DNA hairpins more refined instruments for measuring force. Another key advantage is the fact that their ends are consistently the same distance apart, Salaita says, unlike the random coils of flexible polymers.
In experiments, the DNA hairpins turned out to operate more like a toggle switch than a dimmer switch. "The polymer-based tension probes gradually unwind and become brighter as more force is applied," Salaita says. "In contrast, DNA hairpins don't budge until you apply a certain amount of force. And once that force is applied, they start unzipping and just keep unraveling."
In addition, the researchers were able to calibrate the force constant of the DNA hairpins, making them highly tunable, digital instruments for calculating the amount of force applied by a molecule, down to the piconewton level.
"The force of gravity on an apple is about one newton, so we're talking about a million-millionth of that," Salaita says. "It's sort of mind-bogging that that's how little force you need to unfold a piece of DNA."
The result is a tension probe that is three times more sensitive than the polymer probes.
In a separate paper, published in Nano Letters, the Salaita lab used the DNA-based probes to experiment with how the density of a substrate affects the force applied. "Intuitively you might think that a less dense environment, offering fewer anchoring points, would result in more force per anchor," Salaita said. "We found that it's actually the opposite: You're going to see less force per anchor."
The mechanism of sensing ligand spacing and adhering to a substrate appears to be force-mediated, he says. "The integrin receptors need to be closely spaced in order for the engine in the cell that generates force to engage with them and commit the force."
Now the researchers are using the DNA-based tools they've developed to study the forces of more sensitive cellular pathways and receptors.
"Integrin receptors are kind of beasts, they apply relatively high forces in order to adhere to the extracellular matrix," Salaita says. "There are lots of different cell receptors that apply much weaker forces."
T cells, for example, are white blood cells whose receptors are focused not on adhesion but on activities like distinguishing a friendly self-peptide from a foreign bacterial peptide.
The Salaita lab is collaborating with medical researchers across Emory to understand the role of cellular mechanics in the immune system, blood clotting and neural patterning of axons.
"Basically, our premise is that mechanics play a role in almost all biological processes, and with these DNA-based tension probes we're going to uncover, measure and map those forces," Salaita says.