Jan 18 2016
An innovative method for fast and precise gene sequencing has been proposed by scientists at the National Institute of Standards and Technology (NIST).
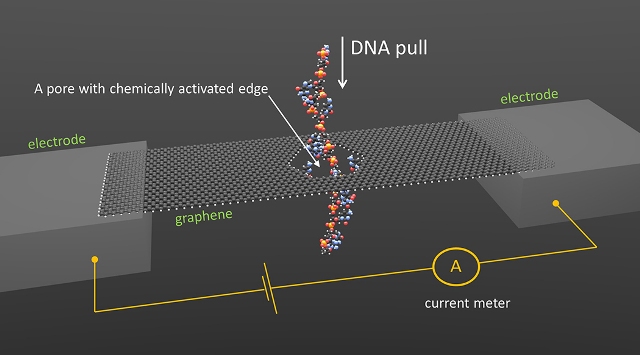 NIST concept for DNA sequencing through a graphene nanopore. Credit: Smolyanitsky/NIST
NIST concept for DNA sequencing through a graphene nanopore. Credit: Smolyanitsky/NIST
The new simulated concept involves pulling a DNA molecule via a small, chemically activated hole in graphene (an ultrathin sheet of carbon atoms), and tracing the changes in the electrical current.
This study may help in detecting around 66 billion bases quickly and easily with 90% accuracy, and without any false positives. Bases are the smallest units of genetic data.
Experimental demonstration of the NIST method may lead to cheaper and faster gene sequencing in comparison to traditional DNA sequencing.
In the 1970s conventional sequencing was developed, this is where parts of DNA are isolated, labeled, copied, and reassembled to read the genetic data. The proposed method provides an alternative to the suggested "nanopore sequencing" concept of pulling DNA via a hole in specific materials. Developed 20 years ago at NIST, this idea is built on the passage of ions or electrically charged particles via the pore. While the idea still remains, it presents a number of issues such as insufficient selectivity and unnecessary electrical noise.
The aim of the proposed method is to produce temporary chemical bonds and depend on graphene's ability to convert the mechanical strains from breaking those bonds into quantifiable blips in electrical current.
This is essentially a tiny strain sensor. We did not invent a complete technology. We outlined a new physical principle that can potentially be far superior to anything else out there.
Alex Smolyanitsky, NIST Theorist
With its compact thin-film structure and unique electrical properties, graphene is increasingly being considered in nanopore-sequencing proposals. In the proposed NIST technique, a graphene nanoribbon measuring 4.5 x 15.5 nm features a number of base copies that are fixed to the nanopore (2.5 nm-wide). The genetic code of the DNA is created from four types of bases. These bases bond as thymine–adenine and cytosine–guanine pairs.
In order to simulate the performance of the sensor in water at room temperature, the NIST team attached the cytosine to the nanopore so as to detect guanine, a nucleobase found in DNA. An unzipped or single-strand DNA molecule is pulled via the pore. As guanine traverses, hydrogen bonds are eventually formed with the cytosine. As the DNA continues to shift, the bonds break, graphene is pulled and subsequently allowed to fall back into position.
The study studied the effect the strain imposed on the electronic properties of graphene, it was observed that temporary changes in electrical current suggest a target base has been passed.
In order to identify all of the four bases, a built-in DNA sensor can be developed. Four graphene ribbons that each have a different base inside the pore, are stacked vertically. To establish the magnitude of quantifiable signal differences, simulated information was combined with hypothesis. Signal strength was found to be in the milliampere range, which is stronger than the one seen in the prior ion-current nanopore techniques. Based on the performance of 90% precision with no false positives, the NIST team proposed that four separate measurements of the same strand of DNA can create 99.99% precision, which is the accuracy needed for human genome sequencing.
The simulated method demonstrates “significant promise for realistic DNA sensing devices” and also eliminates the necessity for sophisticated microscopes, data processing, or limited operating conditions.
Other researchers have experimentally shown the sensor components, as well as fixing the bases to the nanopore. According to the hypothetical analysis, useful electrical signals can be easily separated through the electronic filtering techniques. The NIST method could even be applied with molybdenum disulfide and other strain-sensitive membranes.
A co-author from the University of Groningen in the Netherlands carried out 50% of the simulation, while the NIST team performed the rest of the operation. The paper was titled, “Nucleobase-functionalized graphene nanoribbons for accurate high-speed DNA sequencing”.