Reviewed by Alex SmithNov 25 2021
Scientists in Japan have engineered bottom-up designed peptides for the first time, containing chains of amino acids that can develop synthetic nanopores to identify and facilitate single molecule-sorting of genetic material in a lipid membrane.
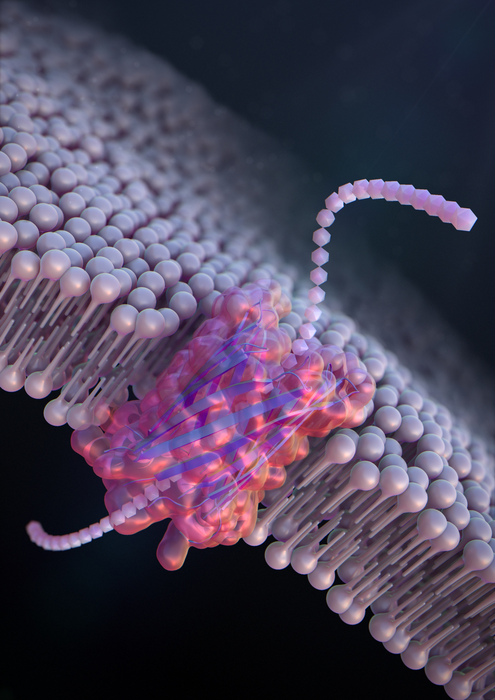 De novo design of a nanopore for single-molecule detection that incorporates a β-hairpin peptide. (Image Credit: Ryuji Kawano, Tokyo University of Agriculture and Technology).
De novo design of a nanopore for single-molecule detection that incorporates a β-hairpin peptide. (Image Credit: Ryuji Kawano, Tokyo University of Agriculture and Technology).
Biological nanopores are large channels composed of pore-forming proteins that can identify specific molecules, but such natural channels are hard to identify, restricting suggested applications in cost-effective, rapid DNA sequencing, small molecule detection, etc.
Nanopore sensing is a powerful tool for label-free, single-molecule detection. This is the first time that DNA and polypeptides were sensed using a de novo-designed nanopore.
Ryuji Kawano, Study Corresponding Author and Professor, Tokyo University of Agriculture and Technology
Their findings have been published in the November 22nd issue of the journal Nature Nanotechnology.
The de novo-engineered nanopores are constructed “from scratch,” according to Kawano, and possess the prospect to mimic natural proteins and their capacity to detect particular proteins.
Significantly, Kawano said, they can also be designed to serve as artificial molecular machines that can detect a much wider variety of molecules—which may help clarify the link between structure and function in target proteins.
Kawano noted that all proteins possessed a novel structure and size.
The folded structure of proteins is determined by their linear polypeptide sequence and gives rise to specific protein functionality. The unique primary structure is the result of structural evolution such as the mutation and selection of amino acid residues over time. To reveal the relationship between this primary information and protein structure is one of the ultimate goals of science.
Ryuji Kawano, Study Corresponding Author and Professor, Tokyo University of Agriculture and Technology
To form large artificial nanopores that can precisely detect and recognize molecules for practical applications, Kawano and the team engineered a peptide called SV28. With two arms of amino acids twisted at a sharp angle, and precise charges at the terminus, the alignment of the hairpin-shaped peptide can be precisely regulated by applying a voltage.
The peptide can orient to form nanopore structures ranging in size from 1.7 to 6.3 nm, ideal for sensing molecules of DNA.
The scientists also altered SV28 by integrating a mutation that causes the peptide structure to bend and twist in particular ways. The resulting peptide developed uniformly dispersed pores of 1.7 nm each, capable of identifying a single polypeptide chain—or one-half of a protein.
This accomplishment could be applied to facilitating the understanding of the link between protein structure and function.
For the subsequent steps, the team aims to design different peptides and proteins to build various types of nanopores to support peptide sequencing, work as molecular robots, and more.
The study’s other contributors are Keisuke Shimizu, Masataka Usami and Ikuro Mizoguchi, Department of Biotechnology and Life Science at Tokyo University of Agriculture and Technology; Batsaikhan Mijiddorj and Izuru Kawamura, Graduate School of Engineering, Yokohama National University; Shuhei Yoshida, Shiori Akayama, Yoshio Hamada and Kenji Usui, Faculty of Frontiers of Innovative Research in Science and Technology, Konan University; and Akifumi Ohyama, Graduate School of Engineering Science, Yokohama National University.
Kawamura is also affiliated with the Graduate School of Engineering Science, Yokohama National University. Mijiddorj is also affiliated with the School of Engineering and Applied Sciences, National University of Mongolia.
This research was supported by the Japan Society for the Promotion of Science (19H05382 and 21H00390), the Ministry of Education, Culture, Sports, Science and Technology (19H00901), and the Mongolian-Japan Engineering Education Development Program partially.
Journal Reference:
Shimizu, K., et al. (2021) De novo design of a nanopore for single-molecule detection that incorporates a β-hairpin peptide. Nature Nanotechnology. doi.org/10.1038/s41565-021-01008-w.